Role of Artificial Intelligence in Drug Discovery and Design: From Foundational Principles to Emerging Applications in Antiviral Therapeutics
Abstract
 Abstract Views: 0
Abstract Views: 0
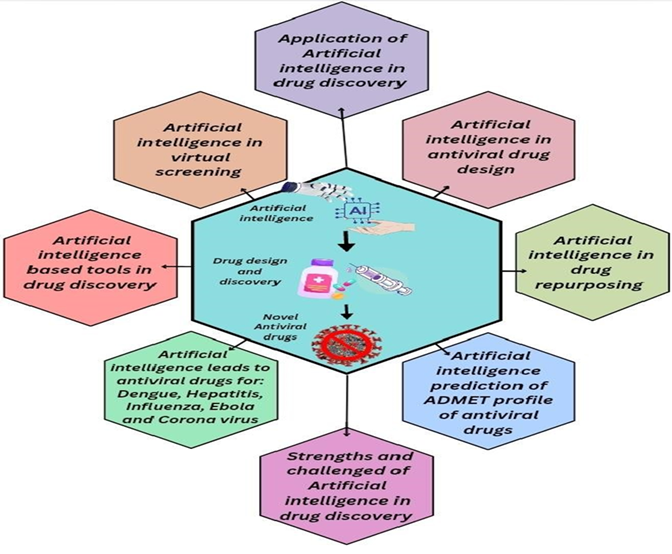
Artificial Intelligence (AI) has significantly transformed drug discovery by enhancing efficiency, reducing costs, and accelerating timelines, particularly in research related to antiviral drugs. Traditional drug discovery methods are not able to compete with rapidly occurring viral mutations, since these are often time-consuming and labor-intensive. Hence, they have been replaced with AI techniques, capable of handling massive datasets, predicting molecular interactions, and optimizing drug candidates rapidly. AI can be used to identify novel drug molecules, drug targets, and repurposed drugs. Furthermore, it can also be used to predict chemical properties, as well as pharmacokinetic, pharmacodynamic, and toxicology profiles by analyzing large datasets. In the early stages of drug discovery, AI aids in target identification and validation by analyzing the genomic, proteomic, and chemical data to predict disease-relevant proteins. In virtual screening and hit identification, AI replaces high-throughput screening with rapid in silico analysis. Generative chemistry approaches utilize reinforcement learning to design novel, drug-like molecules rapidly. Through off-target profiling using models such as DeepTox, AI reduces adverse effects by forecasting unintended protein interactions and drug-drug interactions, improving safety profiles. Its predictive capabilities at each development stage—from molecular screening to clinical trials—have not only accelerated the pace of antiviral drug discovery but have also reduced overall costs significantly, thus proving essential during global pandemics like COVID-19. AI can be implemented at each step of drug discovery and development, from identifying drug molecules and conducting virtual screening to lead optimization and designing clinical trials, as well as interpreting the data obtained from the trials. Antiviral drugs for viral diseases, such as COVID-19, dengue, influenza, hepatitis, and Ebola, developed using AI are mentioned in this study. It also highlights the significance of AI in healthcare, particularly in novel drug development. There is also a dark side to AI, and concerns are rising about the accuracy and quality, as well as the legal and ethical aspects of fact-driven by datasets.
Downloads
References
REFERENCES
Almulhim M, Ghasemian A, Memariani M, et al. Drug repositioning as a promising approach for the eradication of emerging and re-emerging viral agents. Mol Divers. 2025. https://doi.org/10.1007/s11030-025-11131-8
Ozaybi MQB, Madkhali ANM, Alhazmi MAM, et al. The role of artificial intelligence in drug discovery and development. Egypt J Chem. 2024;67(13):1541–1547. https://doi.org/10.21608/ejchem.2024.337877.10835
Namba-Nzanguim CT, Turon G, Simoben CV, et al. Artificial intelligence for antiviral drug discovery in low resourced settings: a perspective. Front Drug Discov. 2022;2:e1013285. https://doi.org/10.3389/fddsv.2022.1013285
Serafim MSM, dos Santos Júnior VS, Gertrudes JC, Maltarollo VG, Honorio KM. Machine learning techniques applied to the drug design and discovery of new antivirals: a brief look over the past decade. Expert Opin Drug Discov. 2021;16(9):961–975. https://doi.org/10.1080/17460441.2021.1918098
Zhou Y, Wang F, Tang J, Nussinov R, Cheng F. Artificial intelligence in COVID-19 drug repurposing. Lancet Dig Health. 2020;2(12):E667–E676. https://doi.org/10.1016/S2589-7500(20)30192-8
Ali M, Park IH, Kim J, et al. How deep learning in antiviral molecular profiling identified anti-SARS-CoV-2 inhibitors. Biomedicines. 2023;11(12):e3134. https://doi.org/10.3390/biomedicines11123134
Liebman M. The role of artificial intelligence in drug discovery and development. Chem Int. 2022;44(1):16–19. https://doi.org/10.1515/ci-2022-0105
Bess A, Berglind F, Mukhopadhyay S, et al. Identification of oral therapeutics using an AI platform against the virus responsible for COVID-19, SARS-CoV-2. Front Pharmacol. 2023;14:e1297924. https://doi.org/10.3389/fphar.2023.1297924
Tarasova O, Poroikov V. Machine learning in discovery of new antivirals and optimization of viral infections therapy. Curr Med Chem. 2021;28(38):7840–7861. https://doi.org/10.2174/0929867328666210504114351
Blanco-Gonzalez A, Cabezon A, Seco-Gonzalez A, et al. The role of AI in drug discovery: challenges, opportunities, and strategies. Pharmaceuticals. 2023;16(6):e891. https://doi.org/10.3390/ph16060891
Arabi AA. Artificial intelligence in drug design: algorithms, applications, challenges and ethics. Future Drug Discov. 2021;3(2):eFDD59. https://doi.org/10.4155/fdd-2020-0028
Nussinov R, Zhang M, Liu Y, Jang H. AlphaFold, artificial intelligence (AI), and allostery. J Phys Chem B. 2022;126(34):6372–6383. https://doi.org/10.1021/acs.jpcb.2c04346
Schaller D, Šribar D, Noonan T, et al. Next generation 3D pharmacophore modeling. Wiley Interdisciplinary Reviews: Comput Mol Sci. 2020;10(4):e1468. https://doi.org/10.1002/wcms.1468
Bhati AP, Wan S, Wright DW, Coveney PV. Rapid, accurate, precise, and reliable relative free energy prediction using ensemble based thermodynamic integration. J Chem Theory Comput. 2017;13(1):210–222. https://doi.org/10.1021/acs.jctc.6b00979
Bunney TD, Wan S, Thiyagarajan N, et al. The effect of mutations on drug sensitivity and kinase activity of fibroblast growth factor receptors: a combined experimental and theoretical study. eBioMedicine. 2015;2(3):194–204. https://doi.org/10.1016/j.ebiom.2015.02.009
Wan S, Knapp B, Wright DW, Deane CM, Coveney PV. Rapid, precise, and reproducible prediction of peptide–MHC binding affinities from molecular dynamics that correlate well with experiment. J Chem Theory Comput. 2015;11(7):3346–3356. https://doi.org/10.1021/acs.jctc.5b00179
Boobier S, Osbourn A, Mitchell JB. Can human experts predict solubility better than computers? J Cheminform. 2017;9:e63. https://doi.org/10.1186/s13321-017-0250-y
Klamt A, Eckert F, Hornig M, Beck ME, Bürger T. Prediction of aqueous solubility of drugs and pesticides with COSMO‐RS. J Comput Chem. 2002;23(2):275–281. https://doi.org/10.1002/jcc.1168
Wu Y, Wang G. Machine learning based toxicity prediction: from chemical structural description to transcriptome analysis. Int J Mol Sci. 2018;19(8):e2358. https://doi.org/10.3390/ijms19082358
Zhao YH, Abraham MH, Ibrahim A, et al. Predicting penetration across the blood-brain barrier from simple descriptors and fragmentation schemes. J Chem Inf Model. 2007;47(1):170–175. https://doi.org/10.1021/ci600312d
Garg P, Verma J. In silico prediction of blood brain barrier permeability: An artificial neural network model. J Chem Inform Model. 2006;46(1):289–297. https://doi.org/10.1021/ci050303i
Rai BK, Bakken GA. Fast and accurate generation of ab initio quality atomic charges using nonparametric statistical regression. J Comput Chem. 2013;34(19):1661–1671. https://doi.org/10.1002/jcc.23308
Olier I, Sadawi N, Bickerton GR, et al. Meta-QSAR: a large-scale application of meta-learning to drug design and discovery. Mach Learn. 2018;107:285–311. https://doi.org/10.1007/s10994-017-5685-x
Tran TTV, Tayara H, Chong KT. Artificial intelligence in drug metabolism and excretion prediction: recent advances, challenges, and future perspectives. Pharmaceutics. 2023;15(4):e1260. https://doi.org/10.3390/pharmaceutics15041260
Tran TTV, Tayara H, Chong KT. Recent studies of artificial intelligence on in silico drug distribution prediction. Int J Mol Sci. 2023;24(3):e1815. https://doi.org/10.3390/ijms24031815
Li B, Tan K, Lao AR, Wang H, Zheng H, Zhang L. A comprehensive review of artificial intelligence for pharmacology research. Front Genet. 2024;15:e1450529. https://doi.org/10.3389/fgene.2024.1450529
Kwofie SK, Adams J, Broni E, et al. Artificial intelligence, machine learning, and big data for Ebola virus drug discovery. Pharmaceuticals. 2023;16(3):e332. https://doi.org/10.3390/ph16030332
Zhang H, Lu XF, Wu Z-P, Lou XWD. Emerging multifunctional single-atom catalysts/nanozymes. ACS Cent Sci. 2020;6(8):1288–1301. https://doi.org/10.1021/acscentsci.0c00512
Lenselink EB, Ten Dijke N, Bongers B, et al. Beyond the hype: deep neural networks outperform established methods using a ChEMBL bioactivity benchmark set. J Cheminform. 2017;9:e45. https://doi.org/10.1186/s13321-017-0232-0
Deng J, Yang Z, Ojima I, Samaras D, Wang F. Artificial intelligence in drug discovery: applications and techniques. Brief Bioinform. 2022;23(1):1–65.
Zhavoronkov A, Ivanenkov YA, Aliper A, et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat Biotechnol. 2019;37(9):1038–1040. https://doi.org/10.1038/s41587-019-0224-x
Pushpakom S, Iorio F, Eyers PA, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. https://doi.org/10.1038/nrd.2018.168
Pan X, Lin X, Cao D, et al. Deep learning for drug repurposing: methods, databases, and applications. Wiley Interdiscipl Rev: Comput Mol Sci. 2022;12(4):e1597. https://doi.org/10.1002/wcms.1597
Mithun R, Shubham JK, Anil GJ. Drug repurposing (DR): an emerging approach in drug discovery. In: Badria FA, ed. Drug Repurposing: Hypothesis, Molecular Aspects and Therapeutic Applications. IntechOpen; 2020:3–20.
Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. https://doi.org/10.1016/j.apsb.2020.02.008
Richardson P, Griffin I, Tucker C, et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395(10223):E30–E31. https://doi.org/10.1016/S0140-6736(20)30304-4
Luo H, Li M, Wang S, Liu Q, Li Y, Wang J. Computational drug repositioning using low-rank matrix approximation and randomized algorithms. Bioinformatics. 2018;34(11):1904–1912. https://doi.org/10.1093/bioinformatics/bty013
Turanli B, Grøtli M, Boren J, et al. Drug repositioning for effective prostate cancer treatment. Front Physiol. 2018;9:e500. https://doi.org/10.3389/fphys.2018.00500
Arora G, Joshi J, Mandal RS, Shrivastava N, Virmani R, Sethi T. Artificial intelligence in surveillance, diagnosis, drug discovery and vaccine development against COVID-19. Pathogens. 2021;10(8):e1048. https://doi.org/10.3390/pathogens10081048
Delijewski M, Haneczok J. AI drug discovery screening for COVID-19 reveals zafirlukast as a repurposing candidate. Med Drug Discov. 2021;9:e100077. https://doi.org/10.1016/j.medidd.2020.100077
Lee MF, Wu YS, Poh CL. Molecular mechanisms of antiviral agents against dengue virus. Viruses. 2023;15(3):e705. https://doi.org/10.3390/v15030705
García-Ariza LL, González-Rivillas N, Díaz-Aguirre CJ, Rocha-Roa C, Padilla-Sanabria L, Castaño-Osorio JC. Antiviral activity of an indole-type compound derived from natural products, identified by virtual screening by interaction on dengue virus NS5 protein. Viruses. 2023;15(7):e1563. https://doi.org/10.3390/v15071563
He X, Zheng Y, Tian C, et al. Quassinoids from Eurycoma longifolia with antiviral activities by inhibiting dengue virus replication. Phytomedicine. 2023;110:e154650. https://doi.org/10.1016/j.phymed.2023.154650
Gautam S, Thakur A, Rajput A, Kumar M. Anti-dengue: a machine learning-assisted prediction of small molecule antivirals against dengue virus and implications in drug repurposing. Viruses. 2023;16(1):e45. https://doi.org/10.3390/v16010045
Moshiri J, Constant DA, Liu B, et al. A targeted computational screen of the SWEETLEAD database reveals FDA-approved compounds with anti-dengue viral activity. mBio. 2020;11(6):e02839-20. https://doi.org/10.1128/mbio.02839-20
Nair V, Chi G, Shu Q, Julander J, Smee DF. A heterocyclic molecule with significant activity against dengue virus. Bioorg Med Chem Lett. 2009;19(5):1425–1427. https://doi.org/10.1016/j.bmcl.2009.01.031
Putri GN, Gudla CS, Singh M, et al. Expanding the anti-flaviviral arsenal: Discovery of a baicalein-derived Compound with potent activity against DENV and ZIKV. Antivir Res. 2023;220:e105739. https://doi.org/10.1016/j.antiviral.2023.105739
Zhou Z, Khaliq M, Suk J-E, et al. Antiviral compounds discovered by virtual screening of small− molecule libraries against dengue virus E protein. ACS Chem Biol. 2008;3(12):765–775. https://doi.org/10.1021/cb800176t
Khalili NSD, Khawory MH, Salin NH, et al. Synthesis and biological activity of imidazole phenazine derivatives as potential inhibitors for NS2B-NS3 dengue protease. Heliyon. 2024;10(2):e24202. https://doi.org/10.1016/j.heliyon.2024.e24202
Bajrai LH, Faizo AA, Alkhaldy AA, Dwivedi VD, Azhar EI. Repositioning of anti-dengue compounds against SARS-CoV-2 as viral polyprotein processing inhibitor. PLoS One. 2022;17(11):e0277328. https://doi.org/10.1371/journal.pone.0277328
World Health Organization. Influenza (seasonal). WHO Web site. https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal). Updated February 28, 2025. Accessed April 20, 2025.
Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2009;457:1012–1014. https://doi.org/10.1038/nature07634
Brownstein JS, Freifeld CC, Madoff LC. Digital disease detection—harnessing the Web for public health surveillance. N Engl J Med. 2009;360(21):2153–2157. https://doi.org/10.1056/NEJMp0900702
Liang G, Zheng L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput Methods Prog Biomed. 2020;187:e104964. https://doi.org/10.1016/j.cmpb.2019.06.023
Pramono RXA, Imtiaz SA, Rodriguez-Villegas E. A cough-based algorithm for automatic diagnosis of pertussis. PLoS One. 2016;11(9):e0162128. https://doi.org/10.1371/journal.pone.0162128
Wang S, Zha Y, Li W, et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur Respir J. 2020;56(2):e2000775. https://doi.org/10.1183/13993003.00775-2020
Heo Y-A. Baloxavir: first global approval. Drugs. 2018;78:693–697. https://doi.org/10.1007/s40265-018-0899-1
Karim MR, Beyan O, Zappa A, et al. Deep learning-based clustering approaches for bioinformatics. Brief Bioinform. 2021;22(1):393–415. https://doi.org/10.1093/bib/bbz170
Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:e14. https://doi.org/10.1038/s41421-020-0153-3
Lim HK, Jeffrey GP, Ramm GA, Soekmadji C. Pathogenesis of viral hepatitis-induced chronic liver disease: role of extracellular vesicles. Front Cell Infect Microbiol. 2020;10:e587628. https://doi.org/10.3389/fcimb.2020.587628
Chen S, Zhang Z, Wang Y, et al. Using quasispecies patterns of hepatitis B virus to predict hepatocellular carcinoma with deep sequencing and machine learning. J Infect Dis. 2021;223(11):1887–1896. https://doi.org/10.1093/infdis/jiaa647
Koirala D, Shao Y, Koldobskaya Y, et al. A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites. Nat Commun. 2019;10:e3629. https://doi.org/10.1038/s41467-019-11585-z
Kanda T, Sasaki R, Masuzaki R, Moriyama M. Artificial intelligence and machine learning could support drug development for hepatitis A virus internal ribosomal entry sites. Artif Intell Gastroenterol. 2021;2(1):1–9. http://dx.doi.org/10.35712/aig.v2.i1.1
Jacob ST, Crozier I, Fischer WA, et al. Ebola virus disease. Nat Rev Dis Primers. 2020;6:e13. https://doi.org/10.1038/s41572-020-0147-3
Emanuel J, Marzi A, Feldmann H. Filoviruses: ecology, molecular biology, and evolution. Adv Virus Res. 2018;100:189–221. https://doi.org/10.1016/bs.aivir.2017.12.002
Muthaiyan M, Naorem LD, Seenappa V, Pushan SS, Venkatesan A. Ebolabase: zaire ebolavirus-human protein interaction database for drug-repurposing. Int J Biol Macromol. 2021;182:1384–1391. https://doi.org/10.1016/j.ijbiomac.2021.04.184
Vamathevan J, Clark D, Czodrowski P, et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18:463–477. https://doi.org/10.1038/s41573-019-0024-5
Brown N, Cambruzzi J, Cox PJ, et al. Big data in drug discovery. Prog Med Chem. 2018;57:277–356. https://doi.org/10.1016/bs.pmch.2017.12.003
Copyright (c) 2025 Nimra Ameer, Saba Razzak, Isha Fatima, Sehrish Haider, Erum Hassan, Amna Baig, Osama Ilyas, Sohail Abbas

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution (CC-BY) 4.0 License that allows others to share the work with an acknowledgement of the work’s authorship and initial publication in this journal









