In Silico Analysis to Predict the Pathogenic Variants of CANT1 Gene Causing Desbuquios Dysplasia (DBQD) Type 1
Abstract
 Abstract Views: 60
Abstract Views: 60
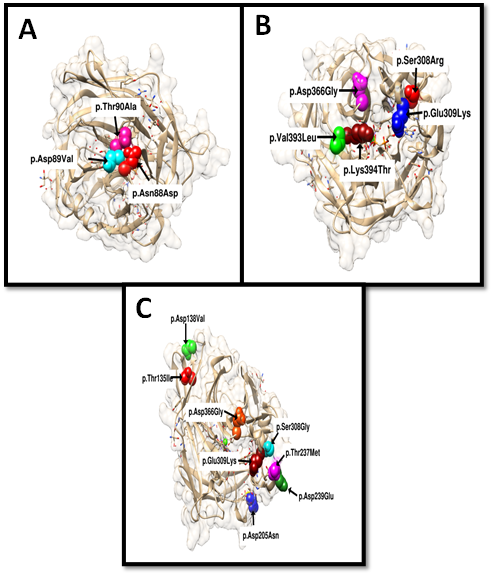
Desbuquois dysplasia (DBQD) is an autosomal recessive chondrodysplasia that belongs to the multiple dislocation group and causes parental and afterbirth growth retardation, hand and proximal femur abnormalities, joint laxity, and scoliosis. Several missense and splice site mutations in CANT1 gene are linked with the development of DBQD. In silico approaches can predict the pathogenic variations causing hereditary diseases. Hence, in the current study, in silico analysis was used to forecast the variants of CANT1 gene that harm the functionality of calcium-dependent nucleotidase. A total of 281 variants with uncertain significance, retrieved from the gnomAD, dbSNP, ClinVar, and Variation Viewer databases, were analyzed using CADD, Meta SNP, CAPiCE, and Condel to predict 61 highly pathogenic variants. Stability change predicting computational tools were applied to filter 19 highly pathogenic amino acid variants that impact protein dynamics via sample conformation or during vibrational entropy. UCSF Chimera was used for interactive visualization and analysis of unwanted interaction among 5 variants in the molecular structure of the protein. Ligand binding computational tools were used to interpret the protein-ligand interactions. A total of three (3) post-translational modification sites were also predictably disrupted by 16 variants. Spice and HSF 3.1 tools were applied to 95 variants to check their disease-causing potential. The variants of the gene were analyzed using computational tools based on different algorithms. The most damaging variants of CANT1 gene that can affect the functionality and stability of the protein were predicted. It was determined that an extensive in silico analysis can determine the likely pathogenic variations for further in vitro experimental analysis.
Downloads
Copyright (c) 2023 Dr. Mureed Husaain

This work is licensed under a Creative Commons Attribution 4.0 International License.
Author(s) retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution (CC-BY) 4.0 License that allows others to share the work with an acknowledgment of the work’s authorship and initial publication in this journal.






