Identification and Analysis of Hub Genes and Biological Pathways Involved in Alzheimer’s Disease (AD) Using Transcriptomics Dataset
Abstract
 Abstract Views: 0
Abstract Views: 0
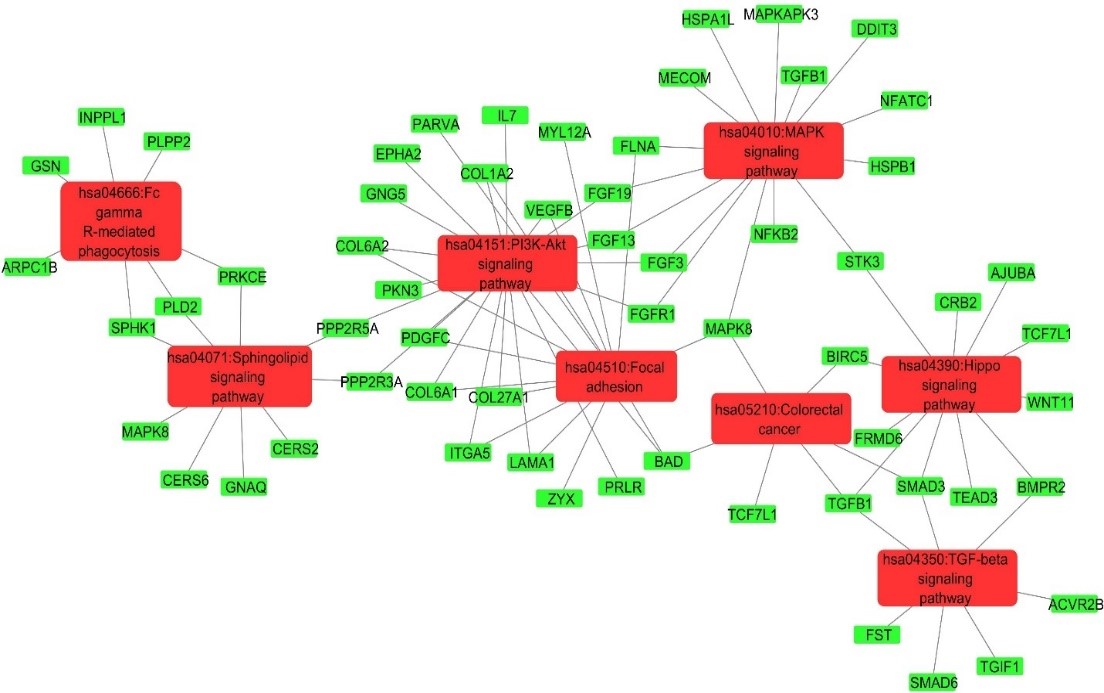
Alzheimer’s disease (AD) is an irreversible and progressive neurodegenerative disorder. The brain mechanisms involved in this disease remain largely unknown. Hence, this study used the integrated bioinformatics approach to analyze a high throughput sequencing dataset (GSE162873) in order to identify the potential biomarkers involved in the pathophysiology of this disease. DESeq2 package was used for the identification of differentially expressed genes (DEGs) from both healthy and diseased patients. DAVID, a web-based bioinformatics resource, was used to perform functional enrichment analysis. StringApp plugin in Cytoscape was utilized to construct the protein-protein interaction (PPI) networks, whereas hub genes were identified through cytoHubba. MCODE was used to perform module analysis, ClueGO to evaluate the KEGG pathways enriched in modules, and miRNet platform for the interaction analysis of miRNAs and hub genes. Drug-genes interaction analysis was performed using DGIdb resource to find out the related drugs. A total of 652 DEGs were screened which were significantly enriched in GO terms. KEGG pathways analysis showed that PI3K-Akt signaling, hippo signaling, MAPK signaling, TGF-beta signaling, and sphingolipid signaling were significantly enriched pathways. A total of 12 hub genes were found to be significantly interacting with miR-603, miR-10b-5p, miR-124-3p, and miR-1-3p, and some FDA approved drugs. The current study provided an insight into the molecular mechanisms of AD and identified some potential biomarker genes, their pathways, miRNAs, and drugs which might be useful for diagnostic and therapeutic purposes.
Downloads
Copyright (c) 2023 Humaira Amin, Asghar Shabbir, Khuram Shahzad

This work is licensed under a Creative Commons Attribution 4.0 International License.
Author(s) retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution (CC-BY) 4.0 License that allows others to share the work with an acknowledgment of the work’s authorship and initial publication in this journal.






