Molecular Dynamics Investigation of Bcl-xL Interactions with Potential Cancer Inhibiting Compounds
Abstract
 Abstract Views: 0
Abstract Views: 0
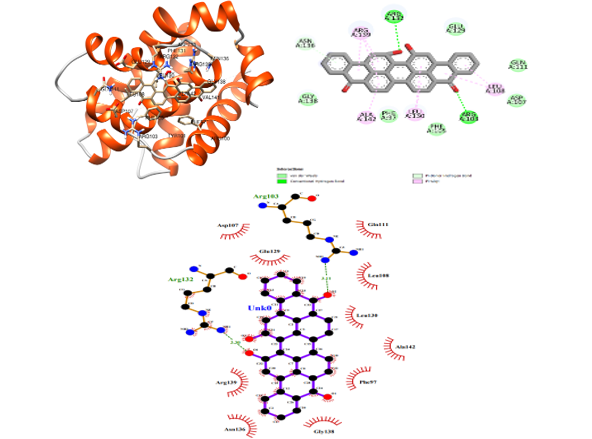
The Bcl-xL protein belongs to the Bcl-2 family of proteins. Bcl-xL is reportedly actively involved in cancer, along with various other factors. Since it has been revealed that this protein plays an active role in cancer, it can be a potential drug target to inhibit cancer activity. Its overexpression can lead to the evasion of apoptosis, allowing cancer cells to persist and proliferate uncontrollably. Hence, targeting Bcl-xL is a promising strategy in cancer therapy, while aiming to induce apoptosis in cancer cells and inhibiting their survival mechanism. In the current study, reported Bcl-xL inhibitors were evaluated for their stability, binding specificity, and interaction dynamics by performing molecular dynamics (MD) simulations. The compounds 24 and 54 revealed stable hydrogen bonding and hydrophobic interactions with active residues in the hydrophobic binding pocket. The binding sites were elucidated for the Bcl-xL protein. Exploring the molecular structural properties involved in binding provides mechanistic insights. This understanding aids in unraveling the protein-ligand interactions crucial for inhibiting anti-apoptotic proteins like Bcl-xL, offering potential for developing targeted inhibitors with anticancer properties.
Downloads
References
Pegoraro L, Palumbo A, Erikson J, et al. A 14; 18 and an 8; 14 chromosome translocation in a cell line derived from an acute B-cell leukemia. Proc Nat Acad Sci. 1984;81(22):7166-7170. https://doi.org/10.1073/pnas.81.22.7166
Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t (14; 18) chromosome translocation. Science. 1984;226(4678):1097-1099. https://doi.org/10.1126/science.6093263
Green DR. The mitochondrial pathway of apoptosis Part II: the BCL-2 protein family. Cold Spring Harbor Perspect Biol. 2022;14(6):e041046.
Ottilie S, Diaz JL, Horne W, et al. Dimerization properties of human Bad: Identification of a BH-3 domain and analysis of its binding to mutant Bcl-2 and Bcl-Xl proteins. J Biol Chem. 1997;272(49):30866-30872. https://doi.org/10.1074/jbc.272.49.30866
Lebedeva I, Rando R, Ojwang J, Cossum P, Stein CA. Bcl-xL in prostate cancer cells: effects of overexpression and down-regulation on chemosensitivity. Cancer Res. 2000;60(21):6052-6060.
Watanabe J, Kushihata F, Honda K, Mominoki K, Matsuda S, Kobayashi N. Bcl-xL overexpression in human hepatocellular carcinoma. Int J Oncol. 2002;21(3):515-519. https://doi.org/10.3892/ijo.21.3.515
Castilla C, Congregado B, Chinchón D, Torrubia FJ, Japón MA, Sáez C. Bcl-xL is overexpressed in hormone-resistant prostate cancer and promotes survival of LNCaP cells via interaction with proapoptotic Bak. Endocrinology. 2006;147(10):4960-4967. https://doi.org/10.1210/en.2006-0502
Noutomi T, Chiba H, Itoh M, Toyota H, Mizuguchi J. Bcl-xL confers multi-drug resistance in several squamous cell carcinoma cell lines. Oral Oncol. 2002;38(1):41-48. https://doi.org/10.1016/S1368-8375(00)00098-1
Chen S, Xing K, Liu S, et al. The role of BCL 2 family proteins in regulating apoptosis and cancer: from mechanistic insights to therapeutic opportunities. Front Oncol. 2022;12:e985363. https://doi.org/10.3389/fonc.2022.985363
Lee EF, Fairlie WD. The structural biology of Bcl-xL. Int J Molecul Sci. 2019;20(9):e2234.
Azam SS, Abro A, Tanvir F, Parvaiz N. Identification of unique binding site and molecular docking studies for structurally diverse Bcl-xL inhibitors. Med Chem Res. 2014;23:3765-3783. https://doi.org/10.1007/s00044-014-0957-5
Borrás C, Mas-Bargues C, Román-Domínguez A, et al. BCL-xL, a mitochondrial protein involved in successful aging: from C. elegans to human centenarians. Int J Molecul Sci. 2020;21(2):e418. https://doi.org/10.3390/ijms21020418
Martin SS, Ridgeway AG, Pinkas J, et al. A cytoskeleton-based functional genetic screen identifies Bcl-xL as an enhancer of metastasis, but not primary tumor growth. Oncogene. 2004;23(26):4641-4645. https://doi.org/10.1038/sj.onc.1207595
Um HD. Bcl-2 family proteins as regulators of cancer cell invasion and metastasis: a review focusing on mitochondrial respiration and reactive oxygen species. Oncotarget. 2016;7(5):e5193. https://doi.org/10.18632/oncotarget.6405
Weiler M, Bähr O, Hohlweg U, et al. BCL-xL: time-dependent dissociation between modulation of apoptosis and invasiveness in human malignant glioma cells. Cell Death Different. 2006;13(7):1156-1169. https://doi.org/10.1038/sj.cdd.4401786
Choi S, Chen Z, Tang LH, et al. Bcl-xL promotes metastasis independent of its anti-apoptotic activity. Nat Commun. 2016;7(1):e10384. https://doi.org/10.1038/ncomms10384
Trisciuoglio D, Tupone MG, Desideri M, et al. BCL-XL overexpression promotes tumor progression-associated properties. Cell Death Dis. 2017;8(12):e3216. https://doi.org/10.1038/s41419-017-0055-y
Dou Z, Zhao D, Chen X, et al. Aberrant Bcl-x splicing in cancer: from molecular mechanism to therapeutic modulation. J Exper Clinical Cancer Res. 2021;40(1):e194. https://doi.org/10.1186/s13046-021-02001-w
Anantram A, Kundaikar H, Degani M, Prabhu A. Molecular dynamic simulations on an inhibitor of anti-apoptotic Bcl-2 proteins for insights into its interaction mechanism for anti-cancer activity. J Biomol Struct Dyn. 2018;37(12):3109-3121. https://doi.org/10.1080/07391102.2018.1508371
Luan J, Hu B, Wang S, et al. Selectivity mechanism of BCL-XL/2 inhibition through in silico investigation. Phy Chem Chem Phy. 2022;24(28):17105-17115. https://doi.org/10.1039/D2CP01755E
Bekker GJ, Fukuda I, Higo J, Fukunishi Y, Kamiya N. Cryptic-site binding mechanism of medium-sized Bcl-xL inhibiting compounds elucidated by McMD-based dynamic docking simulations. Sci Rep. 2021;11(1):e5046. https://doi.org/10.1038/s41598-021-84488-z
Boyenle ID, Ogunlana AT, Oyedele AQ, et al. Reinstating apoptosis using putative Bcl-xL natural product inhibitors: molecular docking and ADMETox profiling investigations. J Taibah Univ Med Sci. 2023;18(3):461-469. https://doi.org/10.1016/j.jtumed.2022.10.014
Nordin N, Khimani K, Abd Ghani MF. Acetogenins exhibit potential BCL-XL Inhibitor for the Induction of apoptosis in the molecular docking study. Current Drug Discoer Technol. 2021;18(6):98-108. https://doi.org/10.2174/1570163818666210204202426
Gunasekaran V, Dhakshinamurthy SS. Computational insights into the interaction of pinostrobin with Bcl-2 family proteins: a molecular docking analysis. Asian Pac J Cancer Prev. 2024;25(2):e507. https://doi.org/10.31557/APJCP.2024.25.2.507
Huang D, Wang Y, Hu B, et al. A computational perspective for tailor-made selective Mcl-1 and Bcl-XL inhibitors. J Mol Struct. 2022;1253:e132269. https://doi.org/10.1016/j.molstruc.2021.132269
Poustforoosh A, Faramarz S, Nematollahi MH, Hashemipour H, Negahdaripour M, Pardakhty A. In silico SELEX screening and statistical analysis of newly designed 5mer peptide-aptamers as Bcl-xl inhibitors using the Taguchi method. Comput Biol Med. 2022;146:e105632. https://doi.org/10.1016/j.compbiomed.2022.105632
Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;9(9):646-652. https://doi.org/10.1038/nsb0902-646
Huey R, Morris GM, Forli S. Using AutoDock 4 and AutoDock vina with AutoDockTools: a tutorial. Scripps Res Inst Molecul Graph Laborat. 2012;10550(92037):e1000.
Mendelsohn LD. ChemDraw 8 ultra, windows and macintosh versions. J Chem Inform Comput Sci. 2004;44(6):2225-2226.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455-461. https://doi.org/10.1002/jcc.21334
Laskowski RA, Swindells MB. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model. 2011:51(10):2778–2786. https://doi.org/10.1021/ci200227u
Studio D. Discovery Studio. Accelrys [2.1]. 2008.
Leontyev IV, Stuchebrukhov AA. Polarizable mean-field model of water for biological simulations with AMBER and CHARMM force fields. J Chem Theory Comput. 2012;8(9):3207-3216. https://doi.org/10.1021/ct300011h
Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:e42717. https://doi.org/10.1038/srep42717
Herschlag D, Pinney MM. Hydrogen bonds: simple after all? Biochemistry. 2018;57(24):3338-3352. https://doi.org/10.1021/acs.biochem.8b00217
Sabri A. Computational modeling of protein dynamics: bridging experimental and theoretical perspectives. Multidiscip J Biochem. 2024;1(1):32-40.
Martínez L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PloS One. 2015;10(3):e0119264. https://doi.org/10.1371/journal.pone.0119264
Gsponer J, Ferrara P, Caflisch A. Flexibility of the murine prion protein and its Asp178Asn mutant investigated by molecular dynamics simulations. J Mol Graph Modell. 2001;20(2):169-82. https://doi.org/10.1016/S1093-3263(01)00117-6
Chakravarty D, Chakraborti S, Chakrabarti P. Flexibility in the N‐terminal actin‐binding domain: clues from in silico mutations and molecular dynamics. Proteins: Struct Funct Bioinf. 2015;83(4):696-710. https://doi.org/10.1002/prot.24767
Zavodszky MI, Kuhn LA. Side‐chain flexibility in protein–ligand binding: the minimal rotation hypothesis. Protein Sci. 2005;14(4):1104-1114. https://doi.org/10.1110/ps.041153605
Fuglebakk E, Echave J, Reuter N. Measuring and comparing structural fluctuation patterns in large protein datasets. Bioinformatics. 2012;28(19):2431-2440. https://doi.org/10.1093/bioinformatics/bts445
Salam AA, Nayek U, Sunil D. Homology modeling and docking studies of Bcl-2 and Bcl-xL with small molecule inhibitors: identification and functional studies. Curr Topics Med Chem. 2018;18(31):2633-2663. https://doi.org/10.2174/1568026619666190119144819
da Silva CP, das Neves GM, Poser GL, Eifler-Lima VL, Rates SM. In silico prediction of ADMET/drug-likeness properties of bioactive phloroglucinols from Hypericum genus. Med Chem. 2023;19(10):1002-1017. https://doi.org/10.2174/1573406419666230601092358
Copyright (c) 2025 Asma Abro, Sumra Wajid Abbasi, Muazma Nabi

This work is licensed under a Creative Commons Attribution 4.0 International License.
Author(s) retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution (CC-BY) 4.0 License that allows others to share the work with an acknowledgment of the work’s authorship and initial publication in this journal.






