Morphological and Genetic Identification of Fish Species of River Ravi, Pakistan
Abstract
 Abstract Views: 0
Abstract Views: 0
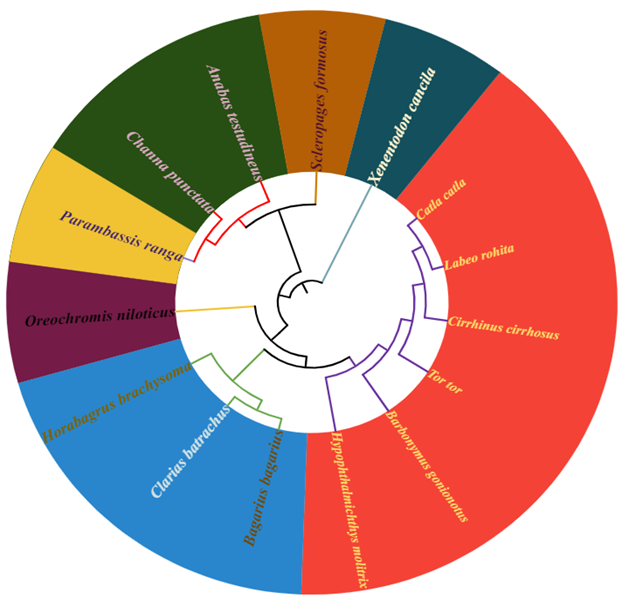
DNA barcoding is a method that examines a specific part of the mitochondrial genome known as the cytochrome c oxidase subunit I (COI) gene, for species identification and biodiversity studies. The current study assessed the effectiveness of using the COI gene for the identification of fish species by constructing a phylogenetic tree with the help of a reference database of fish from the Head Balloki, River Ravi, Pakistan. A total of 15 fish species were analyzed with a 685-base pair (bp) segment of the mitochondrial COI gene sequenced (or "barcoded") for each species. These species represented 15 genera, 10 families, and 6 orders. On average, the sequences were 630 base pairs long. In terms of base composition, the average T content was the highest, while the average G content was the lowest. The AT content (54.1%) was higher than the GC content (45.9%). An analysis of nucleotide pair frequencies across the dataset revealed that out of 685 sites, 385 (56.20%) were conserved within the sequence. Of the 685 sites, 294 sites (42.92%) were variable and 226 (33%) were parsimony informative, while 66 of 685 sites (9.64%) were singletons. In the dataset, transitional pairings (si= 64) were more common than trans-versional pairings (sv = 57), with a si/sv (R) ratio of 1.1. On average, there were 483 pairs where the nucleotides were the same (ii). The average frequencies of nucleotide bases in these sequences were as follows: T (28.8%), C (28.1%), A (25.3%), and G (17.8%). The average Kimura two-parameter (K2P) distances within species, families, and orders were 0.24±0.01%, 0.24±0.02%, and 0.24±0.01%, respectively. Although, each species has a distinct cox1 sequence, various species might occasionally have similar haplotypes. In order to investigate evolutionary links, the maximum likelihood analysis approach was merged with the barcode-based method. In line with the taxonomy categories, the phylogenetic tree generated various groupings that were verified. The current study showed that cox1 sequencing, or DNA barcoding, is a useful method to accurately identify fish species based on both genetic and morphological characteristics.
Downloads
Copyright (c) 2025 Ayesha Eeman, Saima Naz, Ahmad Manan Mustafa Chatha, Syeda Saira Iqbal

This work is licensed under a Creative Commons Attribution 4.0 International License.
Author(s) retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution (CC-BY) 4.0 License that allows others to share the work with an acknowledgment of the work’s authorship and initial publication in this journal.






