In silico Evaluation of the Inhibitory Potential of Novel Hybrid Efflux Pump Inhibitors against Mycobacterium tuberculosis
Abstract
 Abstract Views: 0
Abstract Views: 0
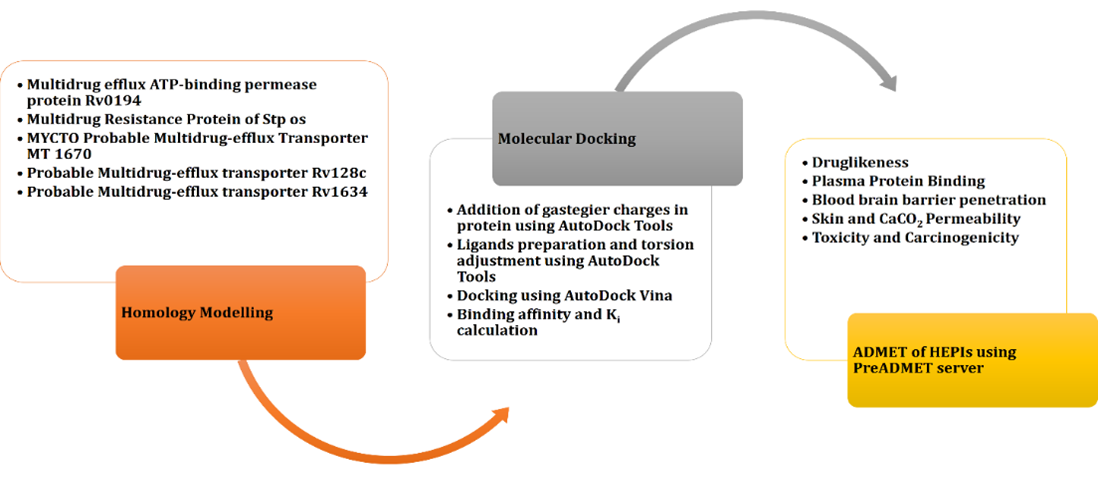
Tuberculosis (TB) is an infectious condition caused by Mycobacterium tuberculosis. One of the key steps towards introducing infectious disease in control is the creation and application of antibiotics. M. tuberculosis strain is multidrug resistant, which is a major threat to TB control. It develops multidrug resistance (MDR) by using efflux pumps (EPs) and other associated systems that can reduce the efficacy of the drug. Several techniques are currently developed to overcome the efflux-mediated resistance and the development of efflux pump inhibitors (EPIs) is one of them. The current study aims to provide an in-silico evaluation of biologically validated hybrid efflux pump inhibitors (HEPIs) with different M. tuberculosis EP proteins. Twelve different HEPIs were identified through literature review. Docking analysis was used to examine the role of HEPI inhibition against 5 MDR EPs. Additionally, the absorption, distribution, metabolism, excretion, and toxicity (ADMET) of all hybrid EP inhibitors were assessed. Molecular docking indicated that several HEPIs, specifically 5, 7, and 11, showed persistent higher binding affinities across multiple proteins, with docking scores comparable to or better than already known inhibitors. The predicted ADMET profiles suggested that most inhibitors had good oral bioavailability and adequate safety margins. To conclude, these HEPIs have the ability to effectively inhibit TB in human beings. In this regard, HEPI-5, HEPI-7, and HEPI-11 were determined as the most promising inhibitors because of their high binding affinities and positive ADMET profiles. Although experimental validation is essential to confirm their therapeutic relevance, these findings highlight their potential as novel EPI scaffolds. However, some of their pharmacological properties are not appropriate for human beings.
Downloads
References
Olmo-Fontánez AM, Turner J. Tuberculosis in an aging world. Pathogens. 2022;11(10):e1101. https://doi.org/10.3390/pathogens11101101
Alland D, Kalkut GE, Moss AR, et al. Transmission of tuberculosis in New York City--an analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;330:1710-1716. https://doi.org/10.1056/NEJM199406163302403
World Health Organization. Global tuberculosis report 2018. WHO Web site. https://www.who.int/publications/i/item/9789241565646
Song Y, Li T, Xia H, et al. Analysis of the epidemiological characteristics of national reported pulmonary tuberculosis incidence, 1997—2023. Chin J Antituberc. 2024;46(10):1198-1208. https://doi.org/10.19982/j.issn.1000-6621.20240382
Falzon D, Schünemann HJ, Harausz E, et al. World Health Organization treatment guidelines for drug-resistant tuberculosis, 2016 update. Eur Respir J. 2017;49(3):e1602308. https://doi.org/10.1183/13993003.02308-2016
Giurazza R, Mazza MC, Andini R, Sansone P, Pace MC, Durante-Mangoni E. Emerging treatment options for multi-drug-resistant bacterial infections. Life. 2021;11(6):e519. https://doi.org/10.3390/life11060519
Alam A, Locher KP. Structure and mechanism of human ABC transporters. Annu Rev Biophys. 2023;52(1):275-300. https://doi.org/10.1146/annurev-biophys-111622-091232
Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuberc Lung Dis. 1998;79(1):3-29. https://doi.org/10.1054/tuld.1998.0002
Belay WY, Getachew M, Tegegne BA, et al. Mechanism of antibacterial resistance, strategies and next-generation antimicrobials to contain antimicrobial resistance: a review. Front Pharmacol. 2024;15:e1444781. https://doi.org/10.3389/fphar.2024.1444781
Huang L, Wu C, Gao H, et al. Bacterial multidrug efflux pumps at the frontline of antimicrobial resistance: an overview. Antibiotics. 2022;11(4):e520. https://doi.org/10.3390/antibiotics11040520
Henderson PJ, Maher C, Elbourne LD, Eijkelkamp BA, Paulsen IT, Hassan KA. Physiological functions of bacterial “multidrug” efflux pumps. Chem Rev. 2021;121(9):5417-5478. https://doi.org/10.1021/acs.chemrev.0c01226
Lake MA, Adams KN, Nie F, et al. The human proton pump inhibitors inhibit Mycobacterium tuberculosis rifampicin efflux and macrophage-induced rifampicin tolerance. Proc Natl Acad Sci. 2023;120(7):e2215512120. https://doi.org/10.1073/pnas.2215512120
Van Bambeke F, Glupczynski Y, Plesiat P, Pechere J, Tulkens PM. Antibiotic efflux pumps in prokaryotic cells: occurrence, impact on resistance and strategies for the future of antimicrobial therapy. J Antimicrob Chemother. 2003;51(5):1055-1065. https://doi.org/10.1093/jac/dkg224
AlMatar M, Albarri O, Makky EA, Köksal F. Efflux pump inhibitors: new updates. Pharmacol Rep. 2021;73:1-16. https://doi.org/10.1007/s43440-020-00160-9
Sreekantan AP, Rajan PP, Mini M, Kumar P. Multidrug efflux pumps in Bacteria and efflux pump inhibitors. Adv Microbiol. 2022;61(3):105-114. https://doi.org/10.2478/am-2022-009
Laws M, Jin P, Rahman KM. Efflux pumps in Mycobacterium tuberculosis and their inhibition to tackle antimicrobial resistance. Trends Microbiol. 2022;30(1):57-68. https://doi.org/10.1016/j.tim.2021.05.001
Pieroni M, Machado D, Azzali E, Costa SS, Couto I, Costantino G, Viveiros M. Rational design and synthesis of thioridazine analogues as enhancers of the antituberculosis therapy. J Med Chem. 2015;58(15):5842-5853. https://doi.org/10.1021/acs.jmedchem.5b00428
Breijyeh Z, Karaman R. Design and synthesis of novel antimicrobial agents. Antibiotics. 2023;12(3):e628. https://doi.org/10.3390/antibiotics12030628
Ch'ng JH, Mok S, Bozdech Z, et al. A whole cell pathway screen reveals seven novel chemosensitizers to combat chloroquine resistant malaria. Sci Rep. 2013;3:e1734. https://doi.org/10.1038/srep01734
Germann UA, Harding MW. Chemosensitizers to overcome and prevent multidrug resistance? Chin Chem Lett. 2025;36(1):e109724. https://doi.org/10.1016/j.cclet.2024.109724
Yu S, Zhang S, Zhang A, Han J, Sun B. Design, Synthesis, and activity evaluation of novel Bifenamide dual-target antibacterial inhibitors and carrier based on infectious microenvironment. J Med Chem. 2025;68(4):4743-4762. https://doi.org/10.1021/acs.jmedchem.4c02913
Asha IJ, Gupta SD, Hossain MM, et al. 2024. In silico characterization of a hypothetical protein (PBJ89160. 1) from Neisseria meningitidis exhibits a new insight on nutritional virulence and molecular docking to uncover a therapeutic target. Evol Bioinform. 2024;20:1-6. https://doi.org/10.1177/11769343241298307
Altunkulah E, Ensari Y. Protein structure prediction: an in-Depth comparison of approaches and tools. Eskişehir Tech Univ J Sci Technol. 2024;13(1):31–51. https://doi.org/10.18036/estubtdc.1378676
Kesheri M, Kanchan S, Häder DP, Sinha RP. Proteomics and bioinformatics approaches for exploring resilience strategies in Cyanobacteria. In: Kesheri M, Kanchan S, Häder DP, Sinha RP, eds. Multi-Omics in Biomedical Sciences and Environmental Sustainability. Springer Nature; 2025:407-427. https://doi.org/10.1007/978-981-96-7067-3_17
Alhusayni AA. The role of AutoDock and related techniques in bioinformatics for developing immune proteins and therapeutic treatments. Infin J Med Innovat. 2025;1(3):62-66.
Tang S, Ding J, Zhu X, et al. Vina-GPU 2.1: towards further optimizing docking speed and precision of AutoDock Vina and its derivatives. IEEE/ACM Transac Comput Biol Bioinfo. 2024;21(6):2382-2393. https://doi.org/10.1109/TCBB.2024.3467127
Nelapati AK, Meena SK. An approach to increase the efficiency of uricase by computational mutagenesis. Phys Chem Res. 2023;11(3):481-491. https://doi.org/10.22036/pcr.2022.345329.2115
Taranto AG, Junior MC. Tools and techniques in structural bioinformatics. In Singh TR, Saini H, Junior MC, eds. Bioinformatics and Computational Biology. Chapman and Hall/CRC; 2023:173-185. https://doi.org/10.1201/9781003331247
Lee SK, Park SH, Lee IH, No KT. PreAD-MET Ver. v2. 0. BMDRC: 2007.
Al-Jarf R, de Sá AG, Pires DE, Ascher DB. pdCSM-cancer: using graph-based signatures to identify small molecules with anticancer properties. J Chem Inf Model. 2021;61(7):3314-3322. https://doi.org/10.1021/acs.jcim.1c00168
Mvondo JG, Matondo A, Mawete DT, Bambi SM, Mbala BM, Lohohola PO. In silico ADME/T properties of quinine derivatives using SwissADME and pkCSM webservers. Int J Trop Dis Health. 2021;42(11):1-2. https://doi.org/10.9734/IJTDH/2021/v42i1130492
Mohammadnabi N, Shamseddin J, Emadi M, et al. Mycobacterium tuberculosis: the mechanism of pathogenicity, immune responses, and diagnostic challenges. J Clin Lab Anal. 2024;38(23):e25122. https://doi.org/10.1002/jcla.25122
Abdel-Halim MS, El-Ganiny AM, Mansour B, Yahya G, Latif HK, Askoura M. Phenotypic, molecular, and in silico characterization of coumarin as carbapenemase inhibitor to fight carbapenem-resistant Klebsiella pneumoniae. BMC Microbiol. 2024;24:e67. https://doi.org/10.1186/s12866-024-03214-7
Zade D. Hepatotoxicity associated with anti-tuberculosis medications: analyzing mechanisms, risk factors, and strategies for prevention and management. J Drug Deliv Biotherap. 2024;1(3):1-12. https://doi.org/10.61920/jddb.v1i03.155
Bertoni Í, Sales BC, Viriato C, Peixoto PV, Pereira LC. Embryotoxicity induced by triclopyr in zebrafish (danio rerio) early life stage. Toxics. 2024;12(4):e255. https://doi.org/10.3390/toxics12040255
Madadi AK, Sohn MJ. Comprehensive therapeutic approaches to tuberculous meningitis: Pharmacokinetics, combined dosing, and advanced intrathecal therapies. Pharmaceutics. 2024;16(4):e540. https://doi.org/10.3390/pharmaceutics16040540
Rodrigues L, Cravo P, Viveiros M. Efflux pump inhibitors as a promising adjunct therapy against drug resistant tuberculosis: a new strategy to revisit mycobacterial targets and repurpose old drugs. Expert Rev Anti Infect Ther. 2020;18(8):741-757. https://doi.org/10.1080/14787210.2020.1760845
Copyright (c) 2025 Ali Raza, Akmal Ali, Siddiqa Batool, Sheikha Rehman, Kazim Raza

This work is licensed under a Creative Commons Attribution 4.0 International License.
Author(s) retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution (CC-BY) 4.0 License that allows others to share the work with an acknowledgment of the work’s authorship and initial publication in this journal.






