Prediction of Breast Cancer Using Machine Learning Techniques
Abstract
 Abstract Views: 497
Abstract Views: 497
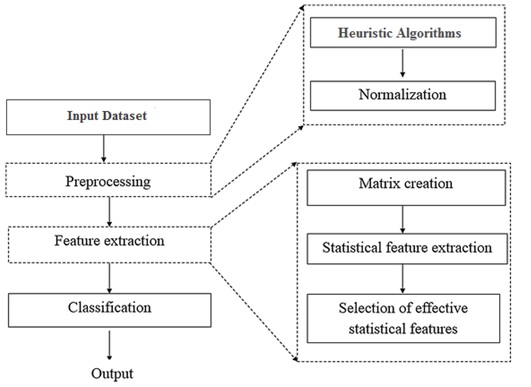
Breast cancer affects a large number of women around the world who are more likely to die as a result of this condition. To seek out the main cause of breast cancer, samples were collected by employing a variety of cutting-edge procedures. The most modern techniques used in this regard are logistic regression, discriminant analysis and principal component analysis (PCA), all of which are useful in determining the causes of breast cancer. The Breast Cancer Wisconsin Diagnostic Dataset collects information about breast cancer via the machine learning repository approach. As a result of the data correlation matrix, we were able to root our job positively. PCA, discriminant analysis, and logistic regression were utilized to extract the dataset features. Models such as decision tree, naive Bayes, logistic regression, support vector machine (SVM), and artificial neural networks were utilized, and their performances were rigorously examined. The results suggested that the proposed strategy works effectively and reduces the training time. These new methods will help doctors to understand the origins of breast cancer and to distinguish between tumor kinds. Data mining techniques are used extensively, especially for feature selection. Finally, it was concluded that among all models, the hybrid discriminant-logistic (DA-LR) feature selection model outperforms SVM and naive Bayes.
Downloads
References
DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics. CA Cancer J Clin. 2013;64(1):52-62. https://doi.org/10.3322/caac.21203
T.A.C. Society. Breast Cancer Early Detection and Diagnosis. [Online]. Available from: https://www.cancer.org/cancer/breast-cancer
. Abe N, Kudo M, Toyama J, Shimbo M. A divergence criterion for classifier-independent feature selection. In Joint IAPR International Workshops on Statistical Techniques in Pattern Recognition (SPR) and Structural and Syntactic Pattern Recognition (SSPR). Springer. Berlin, Heidelberg, 2000;668-676. https://doi.org/10.1007/3 54044522-6_69
. Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Lear Res. 2003;3:1157-1182.
. C Society. Breast Biopsy. [Online]. August 18, 2016. Available from: https://www.cancer.org/cancer/breast cancer/screening-tests-and-early- detection/breastbiopsy.
. Breast Cancer Surveillance Consortium. [Online]. September 23, 2013. Available from: http://www.bcsc-research.org/statistics/performance/screening/2009/rate_age.
. Abdolmaleki P, Buadu LD, Murayama S, Murakami J, Hashiguchi N, Yabuuchi H, Masuda K. Neural network analysis of breast cancer from MRI findings. Radiat Med. 1997;15(5):283-294.
. Abdolmaleki P, Buadu LD, Naderimansh H. Feature extraction and classification of breast cancer on dynamic magnetic resonance imaging using artificial neural network. Cancer Lett. 2001;171(2):183-191. https://doi.org/10.1016/S0304-3835(01)00508-0
. Burke HB, Goodman PH, Rosen DB, et al. Artificial neural networks improve the accuracy of cancer survival prediction. Cancer. 1997;79(4):857-862. https://doi.org/10.1002/(SICI)1097-0142(19970215)79:4<857::AID-CNCR24>3.0.CO;2-Y
. Quinlan JR. Improved use of continuous attributes in C4.5. J Artif Intell Res. 1996;4:77-90.
https://doi.org/10.1613/jair.279
. Pena-Reyes CA, Sipper M. A fuzzy-genetic approach to breast cancer diagnosis. Artif Intell
Med. 1999;17(2):131-155. https://doi.org/10.1016/S0933-3657(99)00019-6
. Hamilton HJ, Cercone N, Shan N. RIAC: a rule induction algorithm based on approximate
classification. University of Regina. 1996.
. Abbass HA. An evolutionary artificial neural networks approach for breast cancer diagnosis. Artif Intell Med. 2002;25(3):265-281. https://doi.org/10.1016/S0933-3657(02)00028-3
. Şahan S, Polat K, Kodaz H, Güneş S. A new hybrid method based on fuzzy-artificial immune system and k-nn algorithm for breast cancer diagnosis. Comput Biol Med. 2007;37(3):415-423. https://doi.org/10.1016/j.compbiomed.2006.05.003
. Akay MF. Support vector machines combined with feature selection for breast cancer diagnosis. Expert Syst Appl. 2009;36(2):3240-3247. https://doi.org/10.1016/j.eswa.2008.01.009
. Chen HL, Yang B, Liu J, Liu DY. A support vector machine classifier with rough set-based feature selection for breast cancer diagnosis. Expert Systems with Applications. 2011;38(7):9014-9022. https://doi.org/10.1016/j.eswa.2011.01.120
. Jin SY, Won JK, Lee H, Choi HJ. Construction of an automated screening system to predict
breast cancer diagnosis and prognosis. Basic Appl Pathol. 2012;5(1):15-18. https://doi.org/10.1111/j.1755-9294.2012.01124.x
. Kaya Y. A new intelligent classifier for breast cancer diagnosis based on a rough set and extreme learning machine: RS+ ELM. Turk J Elec Eng & Comput Sci. 2013;21:2079-2091 https://doi:10.3906/elk-1203-119
. Zheng B, Yoon SW, Lam SS. Breast cancer diagnosis based on feature extraction using a hybrid of K-means and support vector machine algorithms. Expert Syst Appl. 2014;41(4):1476-1482. https://doi.org/10.1016/j.eswa.2013.08.044
. El-Baz AH. Hybrid intelligent system-based rough set and ensemble classifier for breast
cancer diagnosis. Neural Comput Appl. 2015;26(2):437-446.
https://doi.org/10.1007/s00521-014-1731-9
. Bhardwaj A, Tiwari A. Breast cancer diagnosis using genetically optimized neural network
model. Expert Syst Appl. 2015;42(10):4611-4620.
https://doi.org/10.1016/j.eswa.2015.01.065
. Onan A. A fuzzy-rough nearest neighbor classifier combined with consistency-based subset
evaluation and instance selection for automated diagnosis of breast cancer. Exp Syst Appl.
;42(20):6844-6852. https://doi.org/10.1016/j.eswa.2015.05.006
. Örkçü HH, Doğan Mİ, Örkçü M. A Hybrid Applied Optimization Algorithm for Training
MultiLayer Neural Networks in the Data Classification. Gazi Uni J Sci. 2015;28(1):115-
. Aalaei S, Shahraki H, Rowhanimanesh A, Eslami S. Feature selection using genetic
algorithm for breast cancer diagnosis: experiment on three different datasets. Iran J Basic
Med Sci. 2016;19(5):476-482.
. Aličković E, Subasi A. Breast cancer diagnosis using GA feature selection and Rotation
Forest. Neural Comput Appl. 2017;28(4):753-763. https://doi.org/10.1007/s00521-015-
-9
. Yoo I, Alafaireet P, Marinov M, Pena-Hernandez K, Gopidi R, Chang JF, Hua L. Data
mining in healthcare and biomedicine: a survey of the literature. J Med Syst.
;36(4):2431-2448 https://doi.org/10.1007/s10916-011-9710-5 .
. Mitchell TM, Learning M. McGraw-Hill Science. Engineering/Math. 1997;1:27.
. Dey A, Singh J, Singh N. Analysis of Supervised Machine Learning Algorithms for Heart
Disease Prediction with Reduced Number of Attributes using Principal Component
Analysis. Int J Comput Appl. 2016;140(2):27-31.
. Lan K, Wang DT, Fong S, Liu LS, Wong KKL, Dey N. A Survey of Data Mining and Deep
Learning in Bioinformatics. J Med Syst. 2018;42(8):139. https://doi.org/10.1007/s10916-
-1003-9
. Han J, Pei J, Kamber M. Data Mining: Concepts and Techniques. Elsevier. 2011.
. Shiffman D, Fry S, Marsh Z. Cellular Automata. The Nature of Code. 2012:323-330.
. Sharma S, Sharma V, Sharma A. Performance based evaluation of various machine
learning classification techniques for chronic kidney disease diagnosis. Int J Mod Comput
Sci. 2016;4(3):11-16 https://doi.org/10.48550/arXiv.1606.09581
. Peng CY, Lee KL, Ingersoll GM. An introduction to logistic regression analysis and
reporting. J Educ Res. 2002;96(1):3-14. https://doi.org/10.1080/00220670209598786
. Alrashed AA, Gharibdousti MS, Goodarzi M, de Oliveira LR, Safaei MR, Bandarra Filho
EP. Effects on thermophysical properties of carbon based nanofluids: Experimental data,
modelling using regression, ANFIS and ANN. Int J Heat Mass Transf. 2018;125:920-932.
https://doi.org/10.1016/j.ijheatmasstransfer.2018.04.142
. Enders CK. Applied Missing Data Analysis. Methodology in the Social Sciences Series.
Guilford Press. 2010.
. Allison PD. Missing data. Sage Publications. 2001.
. Haitovsky Y. Missing data in regression analysis. J R Stat Soc Series B Stat Methodol.
;30(1):67-82. https://doi.org/10.1111/j.2517-6161.1968.tb01507.x
. Hansen J. Using SPSS for Windows and Macintosh: Analyzing and Understanding Data.
Am Stat.1999;59(1):113-113. https://doi.org/10.1198/tas.2005.s139
. Liong CY, Foo SF. Comparison of linear discriminant analysis and logistic regression for
data classification. InAIP Conference Proceedings. 2013;1522(1):1159-1165.
https://doi.org/10.1063/1.4801262
. Jafari-Marandi R, Davarzani S, Gharibdousti MS, Smith BK. An optimum ANN-based
breast cancer diagnosis: Bridging gaps between ANN learning and decision-making goals.
Appl Soft Comput. 2018;72:108-120. https://doi.org/10.1016/j.asoc.2018.07.060
. Hall MA. Correlation-Based Feature Selection for Machine Learning. [PhD thesis].
Hamilton, New Zealand: The University of Waikato; 1999. Available from:
https://www.cs.waikato.ac.nz/~mhall/thesis.pdf
. Gharibdousti MS, Azimi K, Hathikal S, Won DH. Prediction of chronic kidney disease
using data mining techniques. Proceedings of the 2017 Industrial and Systems Engineering
Conference. 2017;2135-2140.
. Alrashed AA, Gharibdousti MS, Goodarzi M, de Oliveira LR, Safaei MR, Bandarra Filho
EP. Effects on thermophysical properties of carbon based nanofluids: Experimental data,
modelling using regression, ANFIS and ANN. Int J Heat Mass Transf. 2018;125:920-932.
https://doi.org/10.1016/j.ijheatmasstransfer.2018.04.142
. Begdache L, Kianmehr H, Sabounchi N, Chaar M, Marhaba J. Principal component
analysis identifies differential gender-specific dietary patterns that may be linked to mental
distress in human adults. Nutr Neurosci. 2018;23(4):295-308.
https://doi.org/10.1080/1028415X.2018.1500198
Mangal, Anuj, and Vinod Jain. Prediction of Breast Cancer using Machine Learning
Algorithms. Fifth International Conference on I-SMAC (IoT in Social, Mobile, Analytics
and Cloud) (I-SMAC). 2021;464-466. https://doi/10.1109/I-SMAC52330.2021.9640813
Mridha, Krishna. "Early Prediction of Breast Cancer by using Artificial Neural Network
and Machine Learning Techniques. 10th IEEE International Conference on
Communication Systems and Network Technologies (CSNT). 2021;582-587.
Copyright (c) 2022 Tahir Iqbal, Asif Farooq, Nadeem Sarwar, Mohsin Ashraf, Asma Irshad

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










