Identification of Honey Flavonoids as Potential Inhibitors of SARS-CoV-2 RNA-Dependent RNA Polymerase and Main Protease: An In silico Analysis
Abstract
 Abstract Views: 224
Abstract Views: 224
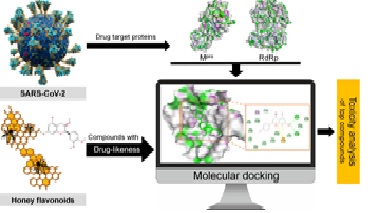
The worldwide spread of COVID-19 caused by SARS-CoV-2 is a serious health crisis which requires a safe and efficacious treatment to combat the disease. RNA-dependent RNA polymerase (RdRp) and main protease (Mpro) are vital enzymes in the life cycle of SARS-CoV-2, considered as effective drug targets. In the current investigation, fourteen (14) honey flavonoids were analyzed for their potential to inhibit RdRp and Mpro using the computational approach. Firstly, flavonoids were screened based on their drug-likeness which determined all the compounds, except epigallocatechin gallate, as orally bioavailable drugs with easy absorbance and high permeability. Secondly, the screened thirteen (13) flavonoids were subjected to molecular docking analysis in order to identify the potent inhibitors of SARS-CoV-2 target proteins (RdRp and Mpro). The analysis revealed significant binding affinity of all compounds with both target proteins. Luteolin showed the most stable binding interaction (−7.6 kcal/mol) with RdRp, while apigenin and kaempferol displayed the binding energy of −7.8 kcal/mol with Mpro. Low binding energies and stable interactions indicated these compounds’ potential inhibition of target proteins. Toxicity analysis depicted these top compounds as safe drugs, which further showed their significant probability to bind the target proteins in human body as a result of target prediction analysis. The above findings predict the anti-COVID-19 potential of honey flavonoids as safe drugs in which top inhibitor compounds exhibit good pharmacodynamic properties and target accuracy. For the future, wet lab experiments involving the in vitro and in vivo assays are recommended to investigate the effectiveness of honey flavonoids in the treatment of COVID-19.
Downloads
References
Peeri NC, Shrestha N, Siddikur Rahman M, et al. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int J Epidemiol. 2021;49(3):717-726. https://doi.org/10.1093/IJE/DYAA033
Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-269. https://doi.org/10.1038/s41586-020-2008-3
Guan W, Ni Z, Hu Y, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382(18):1708-1720. https://doi.org/10.1056/nejmoa2002032
Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497-506. https://doi.org/10.1016/S0140-6736(20)30183-5
Gorbalenya AE, Baker SC, Baric RS, et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536-544. https://doi.org/10.1038/s41564-020-0695-z
Zhou P, Yang X Lou, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270-273. https://doi.org/10.1038/s41586-020-2012-7
Ashour HM, Elkhatib WF, Rahman MM, Elshabrawy HA. Insights into the recent 2019 novel coronavirus (Sars-coV-2) in light of past human coronavirus outbreaks. Pathogens. 2020;9(3):1-15. https://doi.org/10.3390/pathogens9030186
Khan ATA, Khalid Z, Zahid H, Yousaf MA, Shakoori AR. A computational and bioinformatic analysis of ACE2: an elucidation of its dual role in COVID-19 pathology and finding its associated partners as potential therapeutic targets. J Biomol Struct Dyn. 2022;40(4):1813-1829. https://doi.org/10.1080/07391102.2020.1833760
Baric RS, Yount B. Subgenomic Negative-Strand RNA function during mouse hepatitis Virus Infection. J Virol. 2000;74(9):4039-4046. https://doi.org/10.1128/jvi.74.9.4039-4046.2000
Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289-293. https://doi.org/10.1038/s41586-020-2223-y
Liu C, Cai D, Zhang L, et al. Identification of hydrolyzable tannins (punicalagin, punicalin and geraniin) as novel inhibitors of hepatitis B virus covalently closed circular DNA. Antiviral Res. 2016;134. https://doi.org/10.1016/j.antiviral.2016.08.026
Jibril FI, Hilmi ABM, Manivannan L. Isolation and characterization of polyphenols in natural honey for the treatment of human diseases. Bull Natl Res Cent. 2019;43(4). https://doi.org/10.1186/s42269-019-0044-7
Behbahani M. Anti-HIV-1 activity of eight Monofloral Iranian honey types. PLoS One. 2014;9(10):e108195. https://doi.org/10.1371/journal.pone.0108195
Critchfield JW, Butera ST, Folks TM. Inhibition of HIV activation in latently infected cells by flavonoid compounds. AIDS Res Hum Retroviruses. 1996;12(1):39-46. https://doi.org/10.1089/aid.1996.12.39
Fingleton J, Corin A, Sheahan D, et al. Randomised controlled trial of topical kanuka honey for the treatment of cold sores. Adv Integr Med. 2014;1(3):119-123. https://doi.org/10.1016/j.aimed.2014.09.001
Onifade AA, Jewell AP, Ajadi TA, Rahamon SK, Ogunrin OO. Effectiveness of a herbal remedy in six HIV patients in Nigeria. J Herb Med. 2013;3(3):99-103. https://doi.org/10.1016/j.hermed.2013.04.006
Lyu SY, Rhim JY, Park WB. Antiherpetic activities of flavonoids against herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2) in vitro. Arch Pharm Res. 2005;28(11):1293-1301. https://doi.org/10.1007/BF02978215
Chiang LC, Ng LT, Cheng PW, Chiang W, Lin CC. Antiviral activities of extracts and selected pure constituents of Ocimum basilicum. Clin Exp Pharmacol Physiol. 2005;32(10):811-816. https://doi.org/10.1111/j.1440-1681.2005.04270.x
Afrin S, Haneefa SM, Fernandez-Cabezudo MJ, Giampieri F, Al-Ramadi BK, Battino M. Therapeutic and preventive properties of honey and its bioactive compounds in cancer: An evidence-based review. Nutr Res Rev. 2020;33(1):50-76. https://doi.org/10.1017/S0954422419000192
Kim S, Chen J, Cheng T, et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102-D1109. https://doi.org/10.1093/nar/gky1033
Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235-242. https://doi.org/10.1093/nar/28.1.235
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23(1-3):3-25. https://doi.org/10.1016/S0169-409X(96)00423-1
Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1):1-3. https://doi.org/10.1038/srep42717
Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243-250. https://doi.org/10.1007/978-1-4939-2269-7_19
BIOVIA Discovery Studio. Version 4.5. BIOVIA Discovery Studio. https://biovia-discovery-studio-64-bit-client.software.informer.com/4.5/
Pires DEV, Blundell TL, Ascher DB. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066-4072. https://doi.org/10.1021/acs.jmedchem.5b00104n
Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25(2):197-2-6. https://doi.org/10.1038/nbt1284
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357-W364. https://doi.org/10.1093/nar/gkz382
Attique SA, Hassan M, Usman M, et al. A molecular docking approach to evaluate the pharmacological properties of natural and synthetic treatment candidates for use against hypertension. Int J Environ Res Public Health. 2019;16(6):923-923. https://doi.org/10.3390/ijerph16060923
Tsaioun K, Bottlaender M, Mabondzo A. ADDME - Avoiding Drug Development Mistakes Early: Central nervous system drug discovery perspective. BMC Neurol. 2009;9(suppl 1):1-11. https://doi.org/10.1186/1471-2377-9-S1-S1
Talele T, Khedkar S, Rigby A. Successful Applications of Computer Aided Drug Discovery: Moving Drugs from Concept to the Clinic. Curr Top Med Chem. 2010;10(1):127-141. https://doi.org/10.2174/156802610790232251
Murgueitio MS, Bermudez M, Mortier J, Wolber G. In silico virtual screening approaches for anti-viral drug discovery. Drug Discov Today Technol. 2012;9(3):e219-e225. https://doi.org/10.1016/j.ddtec.2012.07.009
Shityakov S, Förster C. In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv Appl Bioinforma Chem. 2014;7(1):23-36. https://doi.org/10.2147/AABC.S63749
Gleeson MP, Hersey A, Montanari D, Overington J. Probing the links between in vitro potency, ADMET and physicochemical parameters. Nat Rev Drug Discov. 2011;10(3):197-208. https://doi.org/10.1038/nrd3367
Duru CE, Duru IA, Adegboyega AE. In silico identification of compounds from Nigella sativa seed oil as potential inhibitors of SARS-CoV-2 targets. Bull Natl Res Cent. 2021;45(1):1-13. https://doi.org/10.1186/s42269-021-00517-x
Mishra A, Rathore AS. RNA dependent RNA polymerase (RdRp) as a drug target for SARS-CoV2. J Biomol Struct Dyn. 2021:1-13. https://doi.org/10.1080/07391102.2021.1875886
Machitani M, Yasukawa M, Nakashima J, Furuichi Y, Masutomi K. RNA-dependent RNA polymerase, RdRP, a promising therapeutic target for cancer and potentially COVID-19. Cancer Sci. 2020;111(11):3976-3976. https://doi.org/10.1111/cas.14618
Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100(2):446-454. https://doi.org/10.1016/j.antiviral.2013.09.015
Shiraki K, Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol Ther. 2020;209:107512-107512. https://doi.org/10.1016/j.pharmthera.2020.107512
Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396). https://doi.org/10.1126/scitranslmed.aal3653
Muralidharan N, Sakthivel R, Velmurugan D, Gromiha MM. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J Biomol Struct Dyn. 2021;39(7):2673-2678. https://doi.org/10.1080/07391102.2020.1752802
Cheke RS. The molecular docking study of potential drug candidates showing anti-covid-19 activity by exploring of therapeutic targets of SARS-CoV-2. Eurasian J Med Oncol. 2020;4(3):185-195. https://doi.org/10.14744/ejmo.2020.31503
Hashem HE. IN Silico approach of some selected honey constituents as SARS-CoV-2 main protease (covid-19) Inhibitors. Eurasian J Med Oncol. 2020;4(3):196-200. https://doi.org/10.14744/ejmo.2020.36102
Zhou J, Fang L, Yang Z, et al. Identification of novel proteolytically inactive mutations in coronavirus 3C-like protease using a combined approach. FASEB J. 2019;33(12):14575-14587. https://doi.org/10.1096/fj.201901624RR
Lv Z, Chu Y, Wang Y. HIV protease inhibitors: A review of molecular selectivity and toxicity. HIV/AIDS - Res Palliat Care. 2015;7:95-95. https://doi.org/10.2147/HIV.S79956
de Leuw P, Stephan C. Protease inhibitors for the treatment of hepatitis C virus infection. GMS Infect Dis. 2017;5:Doc08. https://doi.org/10.3205/id000034
Chiesa N, Rosati B, Arcangeli A, Olivotto M, Wanke E. A novel role for HERG K+ channels: Spike-frequency adaptation. J Physiol. 1997;501(2):313-318. https://doi.org/10.1111/j.1469-7793.1997.313bn.x
Vandenberg JI, Walker BD, Campbell TJ. HERG K+ channels: Friend and foe. Trends Pharmacol Sci. 2001;22(5):240-246. https://doi.org/10.1016/S0165-6147(00)01662-X
Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(W1):W32-W38. https://doi.org/10.1093/nar/gku293
Thomas L, Mathew S, Johnson S. In-silico prediction of role of chitosan, chondroitin sulphate and agar in process of wound healing towards scaffold development. Informatics Med Unlocked. 2020;20:100406-100406. https://doi.org/10.1016/j.imu.2020.100406
Yi L, Li Z, Yuan K, et al. Small Molecules Blocking the Entry of Severe Acute Respiratory Syndrome Coronavirus into Host Cells. J Virol. 2004;78(20):11334-11339. https://doi.org/10.1128/jvi.78.20.11334-11339.2004
Wang S, Ling Y, Yao Y, Zheng G, Chen W. Luteolin inhibits respiratory syncytial virus replication by regulating the MiR-155/SOCS1/STAT1 signaling pathway. Virol J. 2020;17(1):1-4. https://doi.org/10.1186/s12985-020-01451-6
Williamson G, Clifford MN. Colonic metabolites of berry polyphenols: The missing link to biological activity? Br J Nutr. 2010;104:S48-S66. https://doi.org/10.1017/S0007114510003946
Mehla R, Bivalkar-Mehla S, Chauhan A. A flavonoid, luteolin, cripples HIV-1 by abrogation of Tat function. PLoS One. 2011;6(11):e27915. https://doi.org/10.1371/journal.pone.0027915
Fan W, Qian S, Qian P, Li X. Antiviral activity of luteolin against Japanese encephalitis virus. Virus Res. 2016;220:112-116. https://doi.org/10.1016/j.virusres.2016.04.021
Xu L, Su W, Jin J, et al. Identification of luteolin as enterovirus 71 and coxsackievirus A16 inhibitors through reporter viruses and cell viability-based screening. Viruses. 2014;6(7):2778-2795. https://doi.org/10.3390/v6072778
Ryu YB, Jeong HJ, Kim JH, et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorganic Med Chem. 2010;18(22):7940-7947. https://doi.org/10.1016/j.bmc.2010.09.035
Shibata C, Ohno M, Otsuka M, et al. The flavonoid apigenin inhibits hepatitis C virus replication by decreasing mature microRNA122 levels. Virology. 2014;462-463:42-48. https://doi.org/10.1016/j.virol.2014.05.024
Hakobyan A, Arabyan E, Avetisyan A, Abroyan L, Hakobyan L, Zakaryan H. Apigenin inhibits African swine fever virus infection in vitro. Arch Virol. 2016;161(12):3445-3453. https://doi.org/10.1007/s00705-016-3061-y
Wu CC, Fang CY, Cheng YJ, et al. Inhibition of Epstein-Barr virus reactivation by the flavonoid apigenin. J Biomed Sci. 2017;24(1):1-13. https://doi.org/10.1186/s12929-016-0313-9
Wu Q, Yu C, Yan Y, Chen J, Zhang C, Wen X. Antiviral flavonoids from Mosla scabra. Fitoterapia. 2010;81(5):429-433. https://doi.org/10.1016/j.fitote.2009.12.005
Özçelik B, Kartal M, Orhan I. Cytotoxicity, antiviral and antimicrobial activities of alkaloids, flavonoids, and phenolic acids. Pharm Biol. 2011;49(4):396-402. https://doi.org/10.3109/13880209.2010.519390
Lv X, Qiu M, Chen D, Zheng N, Jin Y, Wu Z. Apigenin inhibits enterovirus 71 replication through suppressing viral IRES activity and modulating cellular JNK pathway. Antiviral Res. 2014;109(1):30-41. https://doi.org/10.1016/j.antiviral.2014.06.004
Qian S, Fan W, Qian P, et al. Apigenin restricts FMDV infection and inhibits viral IRES driven translational activity. Viruses. 2015;7(4):1613-1626. https://doi.org/10.3390/v7041613
Schwarz S, Sauter D, Wang K, et al. Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Med. 2014;80(2-3):177-182. https://doi.org/10.1055/s-0033-1360277
Li L, Wang R, Hu H, et al. The antiviral activity of kaempferol against pseudorabies virus in mice. BMC Vet Res. 2021;17(1). https://doi.org/10.1186/s12917-021-02953-3
Behbahani M, Sayedipour S, Pourazar A, Shanehsazzadeh M. In vitro anti-HIV-1 activities of kaempferol and kaempferol-7-Oglucoside isolated from Securigera securidaca. Res Pharm Sci. 2014;9(6):463-469.
Yarmolinsky L, Huleihel M, Zaccai M, Ben-Shabat S. Potent antiviral flavone glycosides from Ficus benjamina leaves. Fitoterapia. 2012;83(2):362-367. https://doi.org/10.1016/j.fitote.2011.11.014
Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;9(9):646-652. https://doi.org/10.1038/nsb0902-646
Copyright (c) 2022 Muhammad Abrar Yousaf

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










