Molecular Epidemiology of SARS-CoV-2 and Correlation of its Clinical Severity with Different Biochemical Parameters: A Retrospective Study
Abstract
 Abstract Views: 0
Abstract Views: 0
Background Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is an infectious disease that was first identified in December 2019 in Wuhan, the capital of China's Hubei province. Since then, it has spread globally, resulting in the ongoing SARS-CoV-2 pandemic. In Pakistan, over 1.5 million cases have been reported since February 2020 (when the first case was reported).
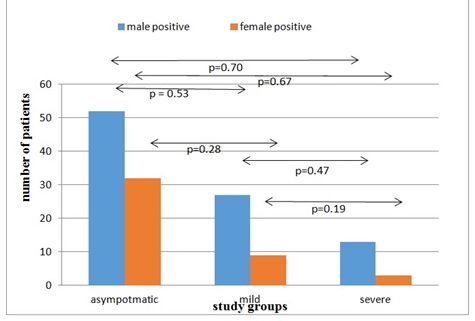
Method This retrospective study was conducted by classifying the data of 136 patients into three study groups, namely asymptomatic (n = 84), mild (n = 36), and severe (n = 16). The data was analyzed using IBM SPSS (version 21).
Results Age and gender showed a non-significant relationship with SARS-CoV-2. Biochemical markers namely D-DIMER, FERRITIN, CRP, and PT showed significant results with p-values 0.001, 0.001, 0.048, and 0.009, respectively (p<0.05). On the contrary, APTT showed a non-significant relationship with SARS-CoV-2 (p= 0.146).
Conclusion It was concluded that the biochemical parameters have seen as the best prediction markers to gauge the SARS-CoV-2 infection severity. Furthermore, this research established the correlation of biochemical parameters with SARS-CoV-2 infection severity and also highlighted the use of these biomarkers as diagnostic and therapeutic biomarkers.
Downloads
References
Giovanetti M, Benedetti F, Campisi G, et al. Evolution patterns of SARS-CoV-2: Snapshot on its genome variants. Biochem Biophys Res Commun. 2021;538:88–91. https://doi. org/10.1016/j.bbrc.2020.10.102
Bivona G, Agnello L, Ciaccio M. Biomarkers for prognosis and treatment response in covid-19 patients. Annals Lab Med. 2021;41(6):540–548. https://doi.org/ 10.3343%2Falm.2021.41.6.540
Wang L, Møhlenberg M, Wang P, Zhou H. Immune evasion of neutralizing antibodies by SARS-CoV-2 Omicron. Cytokine Growth Factor Rev. 2023;70:13–25. https:// doi.org/10.1016/j.cytogfr.2023.03.001
McGowan J, Borucki M, Omairi H, et al. SARS-CoV-2 Monitoring in Wastewater Reveals Novel Variants and Biomarkers of Infection. Viruses. 2022;14(9):e2032. https://doi.org/10.3390/v14092032
Stein SR, Ramelli SC, Grazioli A, et al. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature. 2022;612(7941):758–763. https://doi. org/10.1038/s41586-022-05542-y
de Morais Batista F, Puga MAM, da Silva PV, et al. Serum biomarkers associated with SARS-CoV-2 severity. Sci Rep. 2022;12(1):e15999. https:// doi.org/10.1038/s41598-022-20062-5
Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022;23(1):3–20. https://doi.org/ 10.1038/s41580-021-00418-x
Carabelli AM, Peacock TP, Thorne LG, et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol. 2023;21(3):162–177. https://doi.org/ 10.1038/s41579-022-00841-7
V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. https://doi.org /10.1038/s41579-020-00468-6
Kirtipal N, Bharadwaj S, Kang SG. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect Genet Evol. 2020;85:e104502. https://doi.org/ 10.1016/j.meegid.2020.104502
González-Vázquez LD, Arenas M. Molecular evolution of SARS-CoV-2 during the COVID-19 pandemic. Genes. 2023;14(2);e407 https://doi.org /10.3390/genes14020407
Bai C, Zhong Q, Gao GF. Overview of SARS-CoV-2 genome-encoded proteins. Sci China Life Sci. 2022;65:280–294. https://doi.org/ 10.1007/s11427-021-1964-4
Li S, Liu X, Liu G, Liu C. Biomimetic Nanotechnology for SARS-CoV-2 Treatment. Viruses. 2023;15(3):e596. https://doi.org/10.3390/v15030596
Wang Y, Zhang L, Li Q, et al. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg Microbes Infect. 2022;11(1):1–5. https://doi.org/10. 1080/22221751.2021.2017757
Patil DY, Xu Y, Zhou X, Liu S. Correlation of clinical characteristics between patients with seasonal influenza and patients infected by the wild type or delta variant of SARS-CoV-2. Front Public Health. 2022;10:e981233. https://doi.org/ 10.3389/fpubh.2022.981233
Muralidar S, Ambi SV, Sekaran S, Krishnan UM. The emergence of COVID-19 as a global pandemic: Understanding the epidemiology, immune response and potential therapeutic targets of SARS-CoV-2. Biochimie. 2020;179:85–100. https:// doi.org/10.1016/j.biochi.2020.09.018
Copyright (c) 2024 Maria Rao, Braira Wahid, Syed Sib tul hassan Shah

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










