Mechanisms of Action of Toxins Released by Clostridium perfringens
Abstract
 Abstract Views: 0
Abstract Views: 0
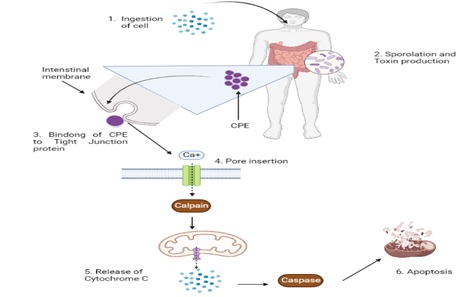
Clostridium perfringens, a rod-shaped anaerobe, is a Gram-positive bacterium that causes foodborne diseases. Its generation time is less than ten minutes and it can divide at 45°C. This aerotolerant bacterium has some toxigenic types (A, B, C, D, and E) that can cause diseases in human beings. Two of its newly discovered toxin types are F and G. Histotoxic, neurological, and intestinal illnesses in both people and animals are instigated by C. perfringens due to its wide range of protein toxins. Alpha or CPA, beta or CPB, epsilon or ETX, iota or ITX, and enterotoxin or CPE are the primary toxins that contribute toward diseases. CPA is the primary pathogenicity factor in gas poisoning in human beings, despite its limited and debatable involvement in animal illnesses. Necrotizing intestinal inflammation and enterotoxaemia in infants of various vertebrate species, particularly humans, are caused by CPB. Some other types cause illnesses in livestock. Necrotic and apoptotic traits are present in the molecular pathways of cell damage linked to C. perfringens toxins.
Downloads
References
Uzal FA, Freedman JC, Shrestha A, et al. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Futr Microbiol. 2014;9(3):361–377. https://doi.org/10.2217/fmb.13.168
Uzal F, Vidal J, McClane B, Gurjar AA. Clostridium perfringens toxins involved in mammalian veterinary diseases. Open Toxicol. 2010;2:24–42.
Garcia J, Beingesser J, Fisher D, et al. The effect of Clostridium perfringens type C strain CN3685 and its isogenic beta toxin null mutant in goats. Vet Microbiol. 2012;157(3-4):412–419. https://doi.org/10.1016/j.vetmic.2012.01.005
Dürre P. From Pandora's Box to Cornucopia: Clostridia–A Historical Perspective. In: Bahl H, Dürre P, eds. Clostridia: Biotechnology and Medical Applications. Wiley Online Library; 2001:1–7. https://doi.org/10.1002/3527600108
Lucey BP, Hutchins GM,William H. Welch MD, and the discovery of Bacillus welchii. Arch Pathol Lab Med. 2004;128(10):1193–1195. https://doi.org/10.5858/2004-128-1193-WHWMAT
Keyburn AL, Bannam TL, Moore RJ, Rood JI. NetB, a pore-forming toxin from necrotic enteritis strains of Clostridium perfringens.Toxins. 2010;2(7):1913–1927. https://doi.org/10.3390/toxins2071913
Stevens DL, Aldape MJ, Bryant AE. Life-threatening clostridial infections. Anaerobe. 2012;18(2):254–259. https://doi.org/10.1016/j.anaerobe.2011.11.001
Labbé RG. Clostridium perfringens. J Assoc Off Anal. Chem. 1991;74(4):711–714.
Fujioka RS, Shizumura LK. Clostridium perfringens, a reliable indicator of stream water quality. J Water Pollut Control Fed. 1985:57(10):986–992.
Nagpal R, Ogata K, Tsuji H, et al. Sensitive quantification of Clostridium perfringens in human feces by quantitative real-time PCR targeting alpha-toxin and enterotoxin genes. BMC Microbiol. 2015;15(1):e219. https://doi.org/10.1186/s12866-015-0561-y
Painter JA, Hoekstra RM, Ayers T, et al. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg Infect Dis. 2013;19(3):407–415. https://doi.org/10.3201%2Feid1903.111866
Tompkins BJ, Wirsing E, Devlin V, et al. Multistate outbreak of Campylobacter jejuni infections associated with undercooked chicken livers—northeastern United States, 2012. MMWR Morb Mortal Wkly Rep. 2013;62(44):874–876.
Kiu R, Caim S, Alexander S, Pachori P, Hall LJ. Probing genomic aspects of the multi-host pathogen Clostridium perfringens reveals significant pangenome diversity, and a diverse array of virulence factors. Front Microbiol. 2017;8:e2485. https://doi.org/10.3389/fmicb.2017.02485
Hatheway CL. Toxigenic clostridia. Clin Microbiol Rev. 1990;3(1):66–98. https://doi.org/10.1128/cmr.3.1.66
Forti K, Ferroni L, Pellegrini M, et al. Molecular characterization of Clostridium perfringens strains isolated in Italy. Toxins. 2020;12(10):e650. https://doi.org/10.3390/toxins12100650
Keyburn AL, Boyce JD, Vaz P, et al. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 2008;4(2):e26. https://doi.org/10.1371/journal.ppat.0040026
Freedman JC, Shrestha A, McClane BA. Clostridium perfringens enterotoxin: action, genetics, and translational applications. Toxins. 2016;8(3):e73. https://doi.org/10.3390/toxins8030073
Grant KA, Kenyon S, Nwafor I, et al. The identification and characterization of Clostridium perfringens by real-time PCR, location of enterotoxin gene, and heat resistance. Foodborne Pathog Dis. 2008;5(5):629–639.https://doi.org/10.1089/fpd.2007.0066
Songer JG. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 1996;9(2):216–234.
Rood JI, McClane BA, Songer JG, Titball RW. The Clostridia: Molecular Biology and Pathogenesis. Academic Press; 1997.
Li J, Ma M, Sarker MR, McClane BA. CodY is a global regulator of virulence-associated properties for Clostridium perfringens type D strain CN3718. MBio. 2013;4(5):e00770–13. https://doi.org/10.1128/mbio.00770-13
Miyamoto K, Yumine N, Mimura K, et al. Identification of novel Clostridium perfringens type E strains that carry an iota toxin plasmid with a functional enterotoxin gene. PLOS ONE. 2011;6(5):e20376. https://doi.org/10.1371/journal.pone.0020376
Lamkanfi M, Dixit VM. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe Rev. 2010;8(1):44–54. https://doi.org/10.1016/j.chom.2010.06.007
Ashida H, Mimuro H, Ogawa M, et al. Cell death and infection: a double-edged sword for host and pathogen survival. J Cell Biol. 2011;195(6):931–942. https://doi.org/10.1083/jcb.201108081
Chaabane W, User SD, El-Gazzah M, et al. Autophagy, apoptosis, mitoptosis and necrosis: interdependence between those pathways and effects on cancer. Arch. Immunol. Ther. Exp. 2013;61:43–58. https://doi.org/10.1007/s00005-012-0205-y
Tait SW, Ichim G, Green DR. Die another way–non-apoptotic mechanisms of cell death. J Cell Sci. 2014;127(10):2135–2144. https://doi.org/10.1242/jcs.093575
Oberst AJ. Death in the fast lane: what's next for necroptosis? FEBS J. 2016;283(14):2616–2625. https://doi.org/10.1111/febs.13520
Buchanan RL. Identification and assessment of exposure to emerging foodborne pathogens using foodborne human viruses as an example. In: Micheal E, Knowles, Lucia E Analick, eds. Present Knowledge in Food Safety. Academic Press; 2023:777–785.
Robertson SL, Smedley III JG, Singh U, et al. Compositional and stoichiometric analysis of Clostridium perfringens enterotoxin complexes in Caco‐2 cells and claudin 4 fibroblast transfectants. Cell Microbiol. 2007;9(11):2734–2755. https://doi.org/10.1111/j.1462-5822.2007.00994.x
Smedley III JG, Uzal FA, McClane BA. Identification of a prepore large-complex stage in the mechanism of action of Clostridium perfringens enterotoxin. Infect Immun. 2007;75(5):2381–2390. https://doi.org/10.1128/iai.01737-06
McClane BA, Uzal FA, Miyakawa MF, et al. The Enterotoxic Clostridia.In: Martin Dworkin, Stanley Falkow, Eugene Rosenberg, Karl-Heinz Schleifer, Erko Stackebrandt, eds. The Prokaryotes. 3rd ed. Springer; 2006;4:698–752. https://doi.org/10.1007/0-387-30744-3_22
Navarro MA, McClane BA, Uzal FA. Mechanisms of action and cell death associated with Clostridium perfringens toxins. Toxins. 2018;10(5):e212. https://doi.org/10.3390/toxins10050212
Chakrabarti G, Zhou X, McClane BA. Death pathways activated in CaCo-2 cells by Clostridium perfringens enterotoxin. Infect Immun. 2003;71(8):4260–4270. https://doi.org/10.1128/iai.71.8.4260-4270.2003
Jewell SA, Titball RW, Huyet J, et al. Clostridium perfringens α-toxin interaction with red cells and model membranes. Soft Matter. 2015;11(39):7748–7761. https://doi.org/10.1039/C5SM00876J
Oda M, Terao Y, Sakurai J, Nagahama MJ. Membrane-binding mechanism of Clostridium perfringens alpha-toxin. Toxins. 2015;7(12):5268–5275. https://doi.org/10.3390/toxins7124880
Flores-Díaz M, Alape-Girón A, Clark G, et al. A cellular deficiency of gangliosides causes hypersensitivity to Clostridium perfringens phospholipase C. J Biol Chem. 2005;280(29):26680–26689. https://doi.org/10.1074/jbc.M500278200
Robert K Y, Tsai YT, Ariga T, Yanagisawa M. Structures, biosynthesis, and functions of gangliosides-an overview. J Oleo Sci. 2011;60(10):537–544. https://doi.org/10.5650/jos.60.537
Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516. https://doi.org/10.1080/01926230701320337
Johansson A-C, Appelqvist H, Nilsson C, Kågedal K, Roberg K, Öllinger KJ. Regulation of apoptosis-associated lysosomal membrane permeabilization. Apoptosis. 2010;15(5):527–540. http://doi.org/10.1007/s10495-009-0452-5
Pinton P, Giorgi C, Siviero R, Zecchini E, Rizzuto R. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene. 2008;27(50):6407–6418. https://doi.org/10.1038/onc.2008.308
Blom T, Slotte JP, Pitson SM, Törnquist KJ. Enhancement of intracellular sphingosine-1-phosphate production by inositol 1, 4, 5-trisphosphate-evoked calcium mobilisation in HEK-293 cells: endogenous sphingosine-1-phosphate as a modulator of the calcium response. Cell Signal. 2005;17(7):827–836. https://doi.org/10.1016/j.cellsig.2004.11.022
Garcia JP, Anderson M, Blanchard P, Mete A, Uzal FA. The pathology of enterotoxemia by Clostridium perfringens type C in calves. J. Vet Diag Invest. 2013;25(3):438–442. https://doi.org/10.1177/1040638713483467
Sayeed S, Uzal FA, Fisher DJ, et al. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol Microbiol. 2008;67(1):15–30. https://doi.org/10.1111/j.1365-2958.2007.06007.x
Uzal FA, Saputo J, Sayeed S, et al. Development and application of new mouse models to study the pathogenesis of Clostridium perfringens type C enterotoxemias. Infect Immun. 2009;77(12):5291–5299. https://doi.org/10.1128/iai.00825-09
Roos S, Wyder M, Candi A, et al. Binding studies on isolated porcine small intestinal mucosa and in vitro toxicity studies reveal lack of effect of C. perfringens beta-toxin on the porcine intestinal epithelium. Toxins. 2015;7(4):1235–1252. https://doi.org/10.3390/toxins7041235
Nagahama M, Ochi S, Oda M, Miyamoto K, Takehara M, Kobayashi KJ. Recent insights into Clostridium perfringens beta-toxin. Toxins 2015;7(2):396–406. https://doi.org/10.3390/toxins7020396
Autheman D, Wyder M, Popoff M, D’herde K, Christen S, Posthaus HJ. Clostridium perfringens beta-toxin induces necrostatin-inhibitable, calpain-dependent necrosis in primary porcine endothelial cells. PLoS One. 2013;8(5):e64644.https://doi.org/10.1371/journal.pone.0064644
Humphries F, Yang S, Wang B, Moynagh PN. RIP kinases: key decision makers in cell death and innate immunity. Cell Death Differ. 2015;22(2):225–236. https://doi.org/10.1038/cdd.2014.126
Popoff MR. Clostridial pore-forming toxins: powerful virulence factors. Anaerobe. 2014;30:220–238. https://doi.org/10.1016/j.anaerobe.2014.05.014
Payne DW, Williamson ED, Havard H, Modi N, Brown J. Evaluation of a new cytotoxicity assay for Clostridium perfringens type D epsilon toxin. FEMS Microbiol Letters. 1994;116(2):161–167. https://doi.org/10.1111/j.1574-6968.1994.tb06695.x
Takagishi T, Oda M, Takehara M, Kobayashi K, Nagahama M. Oligomer formation of Clostridium perfringens epsilon-toxin is induced by activation of neutral sphingomyelinase. Biochim Biophys Acta Biomembr. 2016;1858(11):2681–2688. https://doi.org/10.1016/j.bbamem.2016.07.009
Rumah KR, Ma Y, Linden JR, Oo ML, Anrather J, Schaeren-Wiemers N, Alonso MA, Fischetti VA, McClain MS, Vartanian T. The myelin and lymphocyte protein MAL is required for binding and activity of Clostridium perfringens ε-toxin. PLoS Pathog. 2015;11(5):e1004896. https://doi.org/10.1371/journal.ppat.1004896
Schaeren-Wiemers N, Valenzuela D, Frank M, Schwab MJ. Characterization of a rat gene, rMAL, encoding a protein with four hydrophobic domains in central and peripheral myelin. J Neurosci. 1995;15(8):5753–5764.https://doi.org/10.1523/JNEUROSCI.15-08-05753
Khalili S, Jahangiri A, Hashemi ZS, Khalesi B, Mard-Soltani M, Amani JJ. Structural pierce into molecular mechanism underlying Clostridium perfringens Epsilon toxin function. Toxicon. 2017;127:90–99. https://doi.org/10.1016/j.toxicon.2017.01.010
Petit L, Maier E, Gibert M, Popoff MR, Benz RJ. Clostridium perfringens epsilon toxin induces a rapid change of cell membrane permeability to ions and forms channels in artificial lipid bilayers. J Biol Chem. 2001;276(19):15736–15740. https://doi.org/10.1074/jbc.M010412200
Schmidt G, Papatheodorou P, Aktories KJCoim. Novel receptors for bacterial protein toxins. Curr Opin Microbiol. 2015;23:55–61. https://doi.org/10.1016/j.mib.2014.11.003
Wigelsworth DJ, Ruthel G, Schnell L, et al. CD44 promotes intoxication by the clostridial iota-family toxins. PLoS One. 2012;7(12):e51356. https://doi.org/10.1371/journal.pone.0051356
Tsuge H, Nagahama M, Oda M, et al. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens ι-toxin. Proc Natl Acad Sci. 2008;105(21):7399–7404. https://doi.org/10.1073/pnas.0801215105
Gibert M, Monier MN, Ruez R, et al. Endocytosis and toxicity of clostridial binary toxins depend on a clathrin‐independent pathway regulated by Rho‐GDI. Cell Microbiol. 2011;13(1):154–170. https://doi.org/10.1111/j.1462-5822.2010.01527.x
Nagahama M, Umezaki M, Oda M, et al. Clostridium perfringens iota-toxin b induces rapid cell necrosis. Infect Immun. 2011;79(11):4353–4360. https://doi.org/10.1128/iai.05677-11
Hilger H, Pust S, Von Figura G, et al. The long-lived nature of Clostridium perfringens iota toxin in mammalian cells induces delayed apoptosis. Infect Immun. 2009;77(12):5593–5601. https://doi.org/10.1128/iai.00710-09
Cheng Q, Hwa V, Salyers AA. A locus that contributes to colonization of the intestinal tract by Bacteroides thetaiotaomicron contains a single regulatory gene (chuR) that links two polysaccharide utilization pathways. J Bacteriol. 1992;174(22):7185–7193. https://doi.org/10.1128/jb.174.22.7185-7193.1992
Abraham LJ, Rood JI. Identification of Tn4451 and Tn4452, chloramphenicol resistance transposons from Clostridium perfringens. J Bacteriol. 1987;169(4):1579–1584. https://doi.org/10.1128/jb.169.4.1579-1584.1987
Ridell J, Björkroth J, Eisgrűber H, Schalch B, Stolle A, Korkeala H. Prevalence of the enterotoxin gene and clonality of Clostridium perfringens strains associated with food-poisoning outbreaks. J Food Prot. 1998;61(2):240–243.https://doi.org/10.4315/0362-028X-61.2.240
Fohler S, Klein G, Hoedemaker M, et al. Diversity of Clostridium perfringens toxin-genotypes from dairy farms. BMC Microbiol. 2016;16:e199. https://doi.org/10.1186/s12866-016-0812-6
Copyright (c) 2024 Ali Raza, Sumaira Goshi

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










