Phenotypic Characterization of Multidrug Resistant Klebsiella Spp. Isolated from Pediatric Patients
Abstract
 Abstract Views: 0
Abstract Views: 0
infections are difficult to eradicate due to the presence of β-lactamases. The current study aimed to identify multidrug resistant isolates in children due to their high prevalence in Lahore, Pakistan.
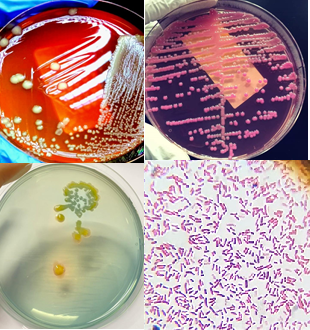
Methodology. In this study, 150 samples of blood, urine, pus, ascitic fluid, pleural fluid, Bronchoalveolar lavage (BAL), and CSF were collected from patients admitted in the University of Child Health Sciences, The Children’s Hospital, Lahore from November 2021 to April 2022. Then, different microbiological techniques including culturing, Gram staining, identification of different species by using API, antibiotic susceptibility testing as well as molecular techniques including PCR, were used to identify MDR Klebsiella spp. positive samples.
Results. Out of the 150 samples, 58 were found positive for Klebsiella spp. while 92 were negative for Klebsiella spp. Among these 58 strains, 48 came from urine samples, 8 from pus samples, and 2 from blood samples. Furthermore, 34 were collected from male and 24 from female patients. The age group of the sampled children was 6 to 10 years. Afterwards, these 58 isolates were screened for extended spectrum β-lactamase (ESBL), metallo β-lactamase (MBL) and AmpC. It was found that out of these 58 isolates, 12 were ESBL positive (20.68%), 11 were MBL positive (18.9%) and 20 were AmpC positive (34.48%). Among β-lactamase genes, NDM -1 (72.7 %), SHV (41.6%), TEM (8.3%) and CTX-M (25%) were detected.
Conclusion. The study concluded that the rate of MDR Klebsiella infection is high in children and it may be complicated for the clinicians to treat them.
Downloads
References
Koirala S, Khadka S, Sapkota S, et al. Prevalence of CTX‐M β‐Lactamases producing multidrug resistant escherichia coli and klebsiella pneumoniae among patients attending Bir hospital, Nepal. BioMed Res Int. 2021;2021(1):e9958294. https://doi.org/10.1155/2021/9958294
Wyres KL, Lam MM, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–359. https://doi.org /10.1038/s41579-019-0315-1
Pal M, Kerorsa GB, Marami LM, Kandi V. Epidemiology, pathogenicity, animal infections, antibiotic resistance, public health significance, and economic impact of staphylococcus aureus: a comprehensive review. Am J Public Health Res. 2020;8(1):14–21.
Yang J, Long H, Hu Y, Feng Y, McNally A, Zong Z. Klebsiella oxytoca complex: update on taxonomy, antimicrobial resistance, and virulence. Clinic Microbiol Rev. 2022;35(1):e00006-21. https:// doi.org/10.1128/CMR.00006-21
Russo A, Fusco P, Morrone HL, Trecarichi EM, Torti C. New advances in management and treatment of multidrug-resistant Klebsiella pneumoniae. Exp Rev Anti-Infect Ther. 2023;21(1):41–55. https://doi.org /10.1080/14787210.2023.2151435
Gharavi MJ, Zarei J, Roshani-Asl P, Yazdanyar Z, Sharif M, Rashidi N. Comprehensive study of antimicrobial susceptibility pattern and extended spectrum beta-lactamase (ESBL) prevalence in bacteria isolated from urine samples. Sci Rep. 2021;11(1):e578. https://doi.org/10. 1038/s41598-020-79791-0
Roy S, Chowdhury G, Mukhopadhyay AK, Dutta S, Basu S. Convergence of biofilm formation and antibiotic resistance in Acinetobacter baumannii infection. Front Med. 202;9:e793615. https://doi.org/10.3389/fmed.2022.793615
Asempa TE, Abdelraouf K, Nicolau DP. Metallo-β-lactamase resistance in Enterobacteriaceae is an artefact of currently utilized antimicrobial susceptibility testing methods. J Antimic Chem. 2020;75(4):997–1005. https://doi.org/10.1093/jac/dkz532
Onduru OG, Mkakosya RS, Aboud S, Rumisha SF. Genetic determinants of resistance among ESBL‐producing enterobacteriaceae in community and hospital settings in east, central, and Southern Africa: a systematic review and meta‐analysis of prevalence. Canad J Infect Dis Med Microbiol. 2021;2021(1):e5153237. https:// doi.org/10.1155/2021/5153237
Tao S, Chen H, Li N, Wang T, Liang W. The spread of antibiotic resistance genes in vivo model. Canad J Infect Dis Med Microbiol. 2022;2022(1):e3348695. https://doi. org/10.1155/2022/3348695
Osman EA, Yokoyama M, Altayb HN, et al. Klebsiella pneumonia in Sudan: multidrug resistance, polyclonal dissemination, and virulence. Antibiotics. 2023;12(2):e233. https:// doi.org/10.3390/antibiotics12020233
Ibrahim ME. Risk factors in acquiring multidrug-resistant Klebsiella pneumoniae infections in a hospital setting in Saudi Arabia. Sci Rep. 2023;13(1):e11626. https://doi.org/10.1038/s41598-023-38871-7
Lam MM, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 202;12(1):e4188. https://doi.org /10.1038/s41467-021-24448-3
Salam MA, Al-Amin MY, Salam MT, et al. Antimicrobial resistance: a growing serious threat for global public health. Healthcare. 2023;11(13):e1946. https://doi.org /10.3390/healthcare11131946
Jian Z, Zeng L, Xu T, et al. Antibiotic resistance genes in bacteria: occurrence, spread, and control. J Basic Microbiol. 2021;61(12):1049–1070. https://doi.org/10.1002/jobm. 202100201
Wood SJ, Kuzel TM, Shafikhani SH. Pseudomonas aeruginosa: infections, animal modeling, and therapeutics. Cells. 2023;12(1):e199. https://doi.org/10.3390/cells12010199
Zhou J, Song S, Xue S, et al. Study of the epidemiological and mechanistic differences between carbapenem-resistant klebsiella pneumoniae infections in children and adults. Infect Drug Resist. 2024;17:2625–2639.
Salawudeen A, Raji YE, Jibo GG, et al. Epidemiology of multidrug-resistant Klebsiella pneumoniae infection in clinical setting in South-Eastern Asia: a systematic review and meta-analysis. Antimicro Resist Infect Cont. 2023;12:e142. https://doi.org/10.1186 /s13756-023-01346-5
Ma J, Gao K, Li M, et al. Epidemiological and molecular characteristics of carbapenem-resistant Klebsiella pneumoniae from pediatric patients in Henan, China. Ann Clinic Microbiol Antimicro. 2024;23:e98. https://doi.org/10.1186/s12941-024-00757-5
Moussa M, Chakra MA, Dellis A, Moussa Y, Papatsoris A. Pharmacotherapeutic advances for recurrent urinary tract infections in women. Exp Opin Pharm. 2020;21(16):2011–2026. https://doi. org/10.1080/14656566.2020.1795128
Junaid K, Ejaz H, Asim I, et al. Heavy metal tolerance trend in extended-spectrum β-lactamase encoding strains recovered from food samples. Int J Environ Res Pub Health. 2021;18(9):e4718. https://doi.org/10. 3390/ijerph18094718
Shahzad S, Willcox MD, Rayamajhee B. A review of resistance to polymyxins and evolving mobile colistin resistance gene (mcr) among pathogens of clinical significance. Antibiotics. 2023:12(11):e1597. https://doi.org/10.3390/antibiotics12111597
Ababu A, Endashaw D, Fesseha H. Isolation and antimicrobial susceptibility profile of Escherichia coli O157: H7 from raw milk of dairy cattle in Holeta District, Central Ethiopia. Int J Microbiol. 2020;2020:e6626488. https://doi.org /10.1155/2020/6626488
Srinivash M, Krishnamoorthi R, Mahalingam PU, et al. Nanomedicine for drug resistant pathogens and COVID-19 using mushroom nanocomposite inspired with bacteriocin–a review. Inorg Chem Commun. 2023;152:e110682. https:// doi.org/10.1016/j.inoche.2023.110682
Janda JM, Abbott SL. The changing face of the family Enterobacteriaceae (Order:“Enterobacterales”): new members, taxonomic issues, geographic expansion, and new diseases and disease syndromes. Clin Microbiol Rev. 2021;34(2):e00174-20. https://doi.org/10.1128/cmr.00174-20
Aslam B, Khurshid M, Arshad MI, et al. Antibiotic resistance: one health one world outlook. Front Cell Infect Microbiol. 2021;11:e771510. https://doi.org/10.3389/fcimb.2021.771510
Copyright (c) 2025 Azka Asghar, Atika Hashmi, Ayesha Khalid, Waqas Ali, Aqsa Ashiq, Abeeha Shahzad Khan

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










