Comparative Analysis of PCR and LIPA Method for HCV Genotypes Screening
Abstract
 Abstract Views: 175
Abstract Views: 175
Abstract
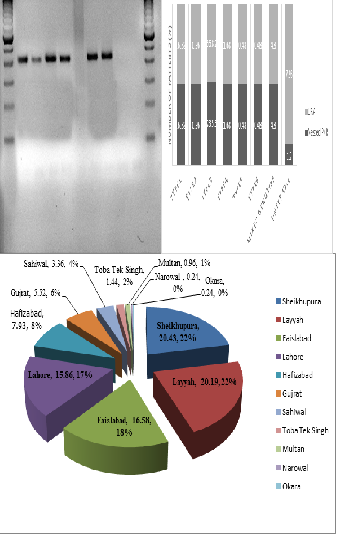
Background: Hepatitis C virus (HCV) is a major health problem worldwide. About 6% of the population of Pakistan is suffering from HCV infection. HCV has a high mutation rate and consists of seven genotypes and sixty-seven subtypes. Genotype information of patients infected with HCV is significant for its treatment.
Methods: In this study, 416 HCV serum samples were collected and HCV prevalence rate was studied in different districts of Punjab, Pakistan. Nested PCR and INNO LIPA HCV-II were used for HCV genotyping and their respective performance was evaluated. This study was conducted by the approval of Lahore Clinical Laboratory and Research Centre situated at Shadman, Lahore.
Results: The highest prevalence of HCV was found in Shekhupura district followed by Bhakkar, Narowal and Okara districts, respectively. In Punjab, the most prevalent genotype was 3a (70.29%), followed by genotype 1 (5.47%), untypable genotypes (5.44%) and genotype 3a/3b (4.64%). Nested PCR was found to be more reliable than INNO LIPA-II. Nested PCR results were more accurate and only 5 samples remained untypable whereas 33 samples could not be typed by LIPA method.
Conclusion: This study was focused on the comparative analysis of Nested PCR and LIPA method for screening HCV genotypes and their prevalence in different districts of Punjab, Pakistan. HCV genotyping is important since different genotypes require different therapeutic treatments. In Punjab, 3a is the most prevalent genotype followed by non-typable genotypes. LIPA is the most commonly used HCV genotype assay but this study found Nested PCR to be a highly sensitive and cost-effective method in this regard. This study can lead to the better selection of genotyping methods and treatment.
Downloads
Copyright (c) 2019 Asma Akhter, Mahwish Javed, Muhammad Imran

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










