High Frequency of Gram-negative Bacilli (GNB) Pathogens in Wounds and Other Clinical Specimens: A Grave Public Health Concern
Abstract
 Abstract Views: 0
Abstract Views: 0
Background. Gram-negative bacilli (GNB) including Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii are important causes of both hospital and community acquired infections in human beings. In this regard, the current study aimed to assess the frequency of GNB pathogens circulating in Hyderabad, Sindh and to obtain locally applicable data for the prevention and spread of infections caused by GNBs.
Methodology. A total of 360 clinical specimens including blood, pus, wound, urine, sputum, and body fluids from suspected indoor and outdoor patients were collected from various diagnostic centers of Hyderabad, Sindh. The isolation, identification, and characterization of GNB pathogens was performed by using standard conventional methods including morphological, cultural, and biochemical testing.
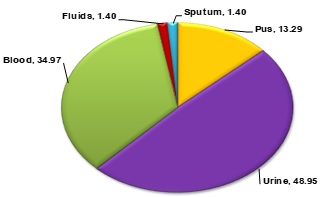
Results. A total of 143 GNBs were isolated and characterized in the current study. The data demonstrated that male patients were more affected with GNBs accounting for 55.94% (n=80) of infected specimens, whereas 44.06% (n=63) of specimens were from female patients. Moreover, specimen wise data of sample positivity revealed that 13.29% (n=19) of GNBs were isolated from pus specimens, 58.59% (n=70) from urine specimens, 34.97% (n=50) from blood specimens, 1.40% (n=2) from fluid specimens, and 1.40% (n=2) of GNBs were isolated from sputum specimens. Bacteriological profiling revealed that 41.26% (n=59) of the isolated bacteria were E. coli, considered as the predominant bacteria isolated from urine specimens. Whereas, S. enterica serover Typhi was the most frequently isolated bacteria from blood specimens accounting for 20.28% (n=29) of all bacteria. Other less prevalent but important pathogenic bacteria included K. pneumoniae accounting for 12.59% (n=18) of all bacteria, P. aeruginosa accounting for 8.39% (n=12) of all bacteria, Acinetobacter spp. accounting for 6.99% (n=10) of all bacteria, and Enterobacter spp. accounting for 2.10% (n=3) of all bacteria.
Conclusion. To conclude, the high frequency of GNBs isolated from clinical specimens at Hyderabad, Sindh poses an alarming situation and warrants an urgent need to monitor and control the spread of pathogenic bacteria.
Downloads
References
Das JK, Hasan R, Zafar A, et al. Trends, Associations, and Antimicrobial Resistance of Salmonella Typhi and Paratyphi in Pakistan. Am J Trop Med Hyg. 2018;99(3 Suppl):48–54. https:// doi.org/10.4269/ajtmh.18-0145.
Greenwald MA & Wolfgang MC. The changing landscape of the cystic fibrosis lung environment: From the perspective of Pseudomonas aeruginosa. Curr Opin Pharmacol. 2022;65:e102262 https://doi.org/10.1016/j.coph.2022.102262
Russo A, Fusco P, Morrone HL, et al. New advances in management and treatment of multidrug-resistant Klebsiella pneumoniae. Expert Rev Anti Infect Ther. 2023;21(1):41–55 https://doi.org/10.1080/14787210.2023.2151435.
Idrees MM, Rimsha R, Idrees MD, et al. Antimicrobial susceptibility and genetic prevalence of extended-spectrum β-lactamases in gram-negative rods isolated from clinical specimens in Pakistan. Antibiotics. 2023;12(1):e29 https://doi.org/10. 3390/antibiotics12010029.
Motbainor H, Bereded F & Mulu W. Multi-drug resistance of blood stream, urinary tract and surgical site nosocomial infections of Acinetobacter baumannii and Pseudomonas aeruginosa among patients hospitalized at Felegehiwot referral hospital, Northwest Ethiopia: a cross-sectional study. BMC Infect Dis. 2020;20(1):e92 https://doi.org/10.1186/s12879-020-4811-8.
Gheysarzadeh A, Pakzad I, Valadbeigi H, et al. Antimicrobial resistance and genetic analysis of multi-drug resistant Klebsiella pneumoniae isolates by pulsed-field gel electrophoresis. Gene Rep. 2020;19:e100638 https://doi.org/10. 1016/j.genrep.2020.100638.
Naha A, Kumar Miryala S, Debroy R, et al. Elucidating the multi-drug resistance mechanism of Enterococcus faecalis V583: A gene interaction network analysis. Gene. 2020;748:e144704 https://doi.org/ 10.1016/j.gene.2020.144704.
Almomani BA, Hayajneh WA, Ayoub AM, et al. Clinical patterns, epidemiology and risk factors of community-acquired urinary tract infection caused by extended-spectrum beta-lactamase producers: a prospective hospital case-control study. Infection. 2018;46(4):495–501 https://doi.org/10.1007/ s15010-018-1148-y.
Kaleem F, Usman J, Hassan A, et al. Frequency and susceptibility pattern of metallo-beta-lactamase producers in a hospital in Pakistan. J Infect Dev Count. 2010;4(12):810–813. https://doi.org/10.3855/jidc.1050.
Oli AN, Eze DE, Gugu TH, et al. Multi-antibiotic resistant extended-spectrum beta-lactamase producing bacteria pose a challenge to the effective treatment of wound and skin infections. Pan Afr Med J. 2017;27:e66 https://doi.org/10. 11604/pamj.2017.27.66.10226.
Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, et al. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev. 2018;31(2):10¬–1128. https://doi.org/10.1128/CMR. 00079-17.
Ruppé É, Woerther P-L, Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Annals Intens Care. 2015;5(1):e21 https://doi.org/10.1186/s13613-015-0061-0.
Lockhart SR, Abramson MA, Beekmann SE, et al. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J Clin Microbiol. 2007; 45(10):3352–3359 https://doi.org/ 10.1128/JCM.01284-07.
Garnacho-Montero J & Amaya-Villar R. The problem of multi-resistance in gram-negative bacilli in intensive care units: Treatment and prevention strategies. Med Intensiva (Engl Ed). 2022;46(6):326–335 https://doi. org/10.1016/j.medine.2022.04.006.
Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12 https://doi.org/ 10.1086/595011.
Paul M, Carrara E, Retamar P, et al. European Society of Clinical Microbiology and Infectious Diseases (ESCMID) guidelines for the treatment of infections caused by multidrug-resistant Gram-negative bacilli (endorsed by European society of intensive care medicine). Clin Microbiol Infect. 2022; 28(4):521–547 https://doi. org/10.1016/j.cmi.2021.11.025.
Odoki M, Almustapha Aliero A, Tibyangye J, et al. Prevalence of bacterial urinary tract infections and associated factors among patients attending hospitals in Bushenyi district, Uganda. Int J Microbiol. 2019;2019:e4246780 https://doi. org/10.1155/2019/4246780.
Iacovelli V, Gaziev G, Topazio L, et al. Nosocomial urinary tract infections: A review. Urologia. 2014;81(4):222–227 https://doi.org /10.5301/uro.5000092.
Ouchar Mahamat O, Lounnas M, Hide M, et al. High prevalence and characterization of extended-spectrum ß-lactamase producing Enterobacteriaceae in Chadian hospitals. BMC Infect Dis. 2019;19:1–7 https://doi.org/ 10.1186/s12879-019-3838-1.
Yadav S, Bhujel R, Hamal P, et al. Burden of Multidrug-Resistant Acinetobacter baumannii Infection in Hospitalized Patients in a Tertiary Care Hospital of Nepal. Infect Drug Resist. 2020;13:725–732. https://doi.org/10.2147/IDR.S239514.
Copyright (c) 2024 Seerat ul Urooj, Shaista Bano, Sarfraz Ali Tunio, Babar Aijaz Memon, Shah Muhammad Abbasi, Zainab Rajput

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










