Investigating Camel Superoxide Dismutase 1: A Computational Analysis of Potential Key Player in Heat Stress Adaptation
Investigating camel superoxide dismutase 1
Abstract
 Abstract Views: 0
Abstract Views: 0
Background. Superoxide dismutase 1 (SOD1) is crucial for cellular defense against oxidative stress induced by superoxide radicals, particularly in challenging conditions, such as elevated temperature and humidity. This study investigated SOD1 in Bactrian camel (Camelus bactrianus), Wild Bactrian camel (C. ferus), and Arabian camel (C. dromedarius) to understand its role in heat tolerance.
Methodology. The current study employed bioinformatics analysis to assess the genomic features including GC% content. It also investigated the structure and location of the SOD1 gene on the chromosomes. Phylogenetic analysis was conducted to elucidate the evolutionary relationships based on SOD1 protein sequences. Structural analyses encompassed secondary and tertiary structure predictions, emphasizing stability and potential functional implications. Subcellular localization of the SOD1 protein was also explored.
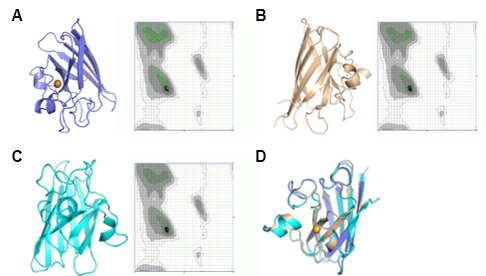
Results. C. dromedarius displayed the highest GC% in its genome, indicating improved thermostability. With the exception of C. bactrianus whose chromosomal location was unknown, all other species contained SOD1 gene on their first chromosome. Based upon SOD1 protein sequences, phylogenetic investigation emphasized the close evolutionary link within the Camelidae family. Structurally, all three species of camel shared an acidic, globular, and thermally-stable SOD1 protein having high glycine content and lack of cleavage sites. Analysis of secondary structure indicated a frequency of random coils, highlighting the adaptability and evolutionary conservation of protein. Predictions of tertiary structure verified that SOD1 was stable in all species. The protein is predominantly found in cytoplasm although, also present in nucleus, extracellular region, and mitochondria.
Conclusion. This inclusive analysis of SOD1 in three different species of camel highlighted their strong adaptation to desert environment by elucidating their genomic and proteomic stability. Further research is necessary to investigate the biochemical mechanisms behind camels’ extraordinary ability to thrive in desert conditions and respond to the challenges posed by climate change.
Downloads
References
Rauf A, Khalil AA, Awadallah S, et al. Reactive oxygen species in biological systems: Pathways, associated diseases, and potential inhibitors—a review. Food Sci Nutr. 2024;12(2):675–693. https://doi.org/10.1002/fsn3.3784
Jena AB, Samal RR, Bhol NK, Duttaroy AK. Cellular Red-Ox system in health and disease: the latest update. Biomed Pharmacother. 2023;162:e114606. https://doi.org/10.1016/j.biopha.2023.114606
Jomova K, Alomar SY, Alwasel SH, Nepovimova E, Kuca K, Valko M. Several lines of antioxidant defense against oxidative stress: antioxidant enzymes, nanomaterials with multiple enzyme-mimicking activities, and low-molecular-weight antioxidants. Arch Toxicol. 2024;98(5):1323–1367. https://doi.org/10.1007/s00204-024-03696-4
Li X, Qiu S, Shi J, et al. A new function of copper zinc superoxide dismutase: as a regulatory DNA-binding protein in gene expression in response to intracellular hydrogen peroxide. Nucleic Acids Res. 2019;47(10):5074–5085. https://doi.org/10.1093/nar/gkz256
Chafik A, Essamadi A, Çelik SY, Mavi A. Purification and biochemical characterization of a novel copper, zinc superoxide dismutase from liver of camel (Camelus dromedarius): an antioxidant enzyme with unique properties. Bioorg Chem. 2019;86:428–436. https://doi.org/10.1016/j.bioorg.2019.02.024
Oselu S, Ebere R, Arimi JM. Camels, Camel milk, and Camel milk product situation in kenya in relation to the world. Int J Food Sci. 2022;2022. https://doi.org/10.1155/2022/1237423
Zakir S, Adem F, Tafese W. Review on composition , nutritive and therapeutic value of camel milk. Int J Innov Sci Res Technol. 2021;6(12):227–233.
Adah A, Ayo J, Adah D. Unique physiological and behavioural adaptive features of the one-humped Camel (Camelus dromedarius) to arid environments. J Appl Vet Sci. 2022;8(1):57–64.
Padalino B, Menchetti L. The first protocol for assessing the welfare of dromedary camels (Camelus dromedarius) kept under nomadic pastoralism. Front Vet Sci. 2024;11:e1416714. https://doi.org/10.3389/fvets.2024.1416714
Demirci-Çekiç S, Özkan G, Avan AN, Uzunboy S, Çapanoğlu E, Apak R. Biomarkers of oxidative stress and antioxidant defense. J Pharm Biomed Anal. 2022;209:e114477.
Khan ZA, Mishra C, Jyotiranjan T. In silico analysis of caprine superoxide dismutase 1 (SOD1) gene. Genomics. 2020;112(1):212–217. https://doi.org/10.1016/j.ygeno.2019.01.016
Bateman A, Martin MJ, Orchard S, et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489.
Garg VK, Avashthi H, Tiwari A, et al. MFPPI – multi FASTA protparam Interface. Bioinformation. 2016;12(2):74–77. https://doi.org/10.6026/97320630012074
Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–971. https://doi.org/10.1038/nprot.2007.131
Geourjon C, Deléage G. Sopma: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics. 1995;11(6):681–684.
Burley SK, Bhikadiya C, Bi C, et al. Protein data bank: celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D. Protein Sci. 2022;31(1):187–208. https://doi.org/10.1002/pro.4213
Robin X, Waterhouse AM, Bienert S, et al. The SWISS-MODEL repository of 3D protein structures and models. In: Daina A, Przewosny M, Zoete V, eds. Open Access Databases and Datasets for Drug Discovery. Wiley Publishers; 2024:175–99.
Abramson J, Adler J, Dunger J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630(8016):493–500. https://doi.org/10.1038/s41586-024-07487-w
Powell HR, Islam SA, David A, Sternberg MJE. Phyre2.2: a community resource for template-based protein structure prediction. J Mol Biol. 2025;e168960. https://doi.org/10.1016/j.jmb.2025.168960
Yu CS, Cheng CW, Su WC, et al. CELLO2GO: a web server for protein subCELlular lOcalization prediction with functional gene ontology annotation. PLoS One. 2014;9(6):e99368. https://doi.org/10.1371/journal.pone.0099368
Raza SHA, Hassanin AA, Dhshan AIM, et al. In silico genomic and proteomic analyses of three heat shock proteins (HSP70, HSP90-α, and HSP90-β) in even-toed ungulates. Electron J Biotechnol. 2021;53:61–70. https://doi.org/10.1016/j.ejbt.2021.07.002
Chen H, Skylaris CK. Analysis of DNA interactions and GC content with energy decomposition in large-scale quantum mechanical calculations. Phys Chem Chem Phys. 2021;23(14):8891–8899. https://doi.org/10.1039/D0CP06630C
Kebir NE, Berber N, Zahzeh MR. Anatomical and physiological properties of the dromedary: a potential sustainability alternative and a vital asset in the era of climate change. J Anim Behav Biometeorol. 2024;12(4):e2024031. https://doi.org/10.31893/jabb.2024031
Bornstein S. Evolution, distribution, and economic importance of the Camels. In: Khalafalla AI, Hussein MF, eds. Infectious Diseases of Dromedary Camels: A Concise Guide. Springer International Publishing; 2021:1–19.
Qing R, Hao S, Smorodina E, Jin D, Zalevsky A, Zhang S. Protein design: from the aspect of water solubility and stability. Chem Rev. 2022;122(18):14085–14179.
Gamage DG, Gunaratne A, Periyannan GR, Russell TG. Applicability of instability index for in vitro protein stability prediction. Protein Pept Lett. 2019;26(5):339–347. https://doi.org/10.2174/0929866526666190228144219
Flores-Castañón N, Sarkar S, Banerjee A. Structural, functional, and molecular docking analyses of microbial cutinase enzymes against polyurethane monomers. J Hazard Mater Lett. 2022;3:e100063. https://doi.org/10.1016/j.hazl.2022.100063
Grossmann L, McClements DJ. Current insights into protein solubility: a review of its importance for alternative proteins. Food Hydrocoll. 2023;137:e108416. https://doi.org/10.1016/j.foodhyd.2022.108416
Kovács D, Bodor A. The influence of random-coil chemical shifts on the assessment of structural propensities in folded proteins and IDPs. RSC Adv. 2023;13(15):10182–10203. https://doi.org/10.1039/D3RA00977G
Guan T, Guo Y, Zhou T, et al. Oxidized SOD1 accelerates cellular senescence in neural stem cells. Stem Cell Res Ther. 2024;15(1):e55. https://doi.org/10.1186/s13287-024-03669-5
Martinelli I, Zucchi E, Simonini C, et al. The landscape of cognitive impairment in superoxide dismutase 1-amyotrophic lateral sclerosis. Neural Regen Res. 2023;18(7):1427–1433. https://doi.org/10.4103/1673-5374.361535
Copyright (c) 2025 Muhammad Abrar Yousaf, Muhammad Asjad Khn

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










