In Silico Characterization of Hypothetical Protein AZJ53_10480 in Streptococcus pneumoniae
Abstract
 Abstract Views: 0
Abstract Views: 0
Background. Streptococcus pneumoniae is a major human pathogen responsible for serious infections such as pneumonia. Despite extensive research, many proteins in S. pneumoniae, including hypothetical proteins, remain uncharacterized, limiting the understanding of the bacterium's pathogenic mechanisms.
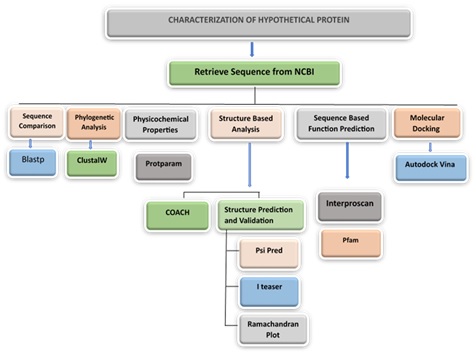
Methods. This study utilizes in silico tools to characterize the hypothetical protein AZJ53_10480 from S. pneumoniae. Sequence alignment and phylogenetic analysis were conducted using BLASTp and ClustalW, while PSIPRED and I-TASSER predicted the protein’s secondary and tertiary structures. Molecular docking studies were performed with AutoDock Vina to assess potential interactions with the antiviral drug sofosbuvir
Results. The in silico analysis revealed that the hypothetical protein AZJ53_10480 shares structural and functional similarities with viral capsid proteins of the hepatitis C virus. The protein was found to have a mixed localization, suggesting potential multifunctionality within the bacterial cell. Molecular docking studies indicated a strong binding affinity between AZJ53_10480 and sofosbuvir, suggesting that this protein could be a potential target for therapeutic intervention.
Conclusion. This study highlights structural properties and functional roles of hypothetical protein AZJ53_10480 in S. pneumoniae of . The findings suggest that AZJ53_10480 may play a role in the pathogenicity of this bacterium and could serve as a novel target for therapeutic development. Further experimental studies are needed to validate these findings and explore the protein's potential as a drug target.
Highlights
- In silico analysis reveals structural similarities between the hypothetical protein AZJ53_10480 and viral capsid proteins.
- Docking studies suggest AZJ53_10480 as a potential target for antiviral drugs like sofosbuvir.
- The study provides new insights into the possible pathogenic role of hypothetical proteins in Streptococcus pneumoniae.
Downloads
References
Appelbaum PC. Antimicrobial resistance in streptococcus pneumoniae: an overview. Clinic Infect Dis. 1992:15(1):77–83. https://doi.org/10.1093/clinids/15.1.77
Lynch JP, Zhanel GG. Streptococcus pneumoniae: epidemiology, risk factors, and strategies for prevention. Semin Respir Crit Care Med. 2009;30(2):189–209. https://doi.org /10.1055/s-0029-1202938
Rabbi MF, Akter SA, Hasan MJ, Amin A. In silico characterization of a hypothetical protein from Shigella dysenteriae ATCC 12039 reveals a pathogenesis-related protein of the type-VI secretion system. Bioinf Biol Insights. 2021;15:1–12. https://doi.org /10.1177/11779322211011140
Ahmed F, Ahmed N, Prome AA, Robin TB, Rani NA. In silico identification of Shigella sonnei hypothetical protein RUK71877. 1 as interleukin receptor mimic Protein A and a potential drug target. Int J Biosci. 2022;21(6);7–17.
Tasneem M, Gupta SD, Momin MB, Hossain KM, Osman TB, Rabbi MF. In silico annotation of a hypothetical protein from Listeria monocytogenes EGD-e unfolds a toxin protein of the type II secretion system. Genom Inform. 2023;21(1):e7. https://doi.org /10.5808%2Fgi.22071
Shahrear S, Zinnia MA, Sany MR, Islam AB. Functional analysis of hypothetical proteins of vibrio parahaemolyticus reveals the presence of virulence factors and growth-related enzymes with therapeutic potential. Bioinf Biol Insights. 2022;16:1–16. https://doi.org/10.1177/11779322221136002
Rabbi MF, Akter SA, Hasan MJ, Amin A. In silico characterization of a hypothetical protein from ATCC 12039 reveals a pathogenesis-related protein of the type-VI secretion system. Bioinf Biol Insights. 2021;15:1–12. https://doi.org/10.1177/ 11779322211011140
Masum MH, Rajia S, Bristi UP, et al. In silico functional characterization of a hypothetical protein from pasteurella multocida reveals a novel s-adenosylmethionine-dependent methyltransferase activity. Bioinf Biol Insights. 2023;17:1–17. https://doi.org /10.1177/11779322231184024
Munna MM, Islam MA, Shanta SS, Monty MA. Structural, functional, molecular docking analysis of a hypothetical protein from Talaromyces marneffei and its molecular dynamic simulation: an in-silico approach. J Biomol Struc Dyn. 2024:1–20. https://doi.org/10.1080/07391102.2024.2314264
Morris GM, Lim-Wilby M. Molecular docking. In: Kukol A, ed. Molecular Modeling of Proteins. Springer; 2008:365–382. https://doi.org/10. 1007/978-1-59745-177-2_19
McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. https://doi.org /10.1093/bioinformatics/16.4.404
Liu J, Rost B. Sequence-based prediction of protein domains. Nucl Acids Res. 2004;32(12):3522–3530. https://doi.org/10.1093/nar/gkh684
Mondol SM, Das D, Priom DM, Rahman MS, Islam MR, Rahaman MM. In silico identification and characterization of a hypothetical protein from rhodobacter capsulatus revealing s-adenosylmethionine-dependent methyltransferase activity. Bioinf Biol Insights. 2022;16:1–16. https://doi.org/10.1177/11779322221094236
Laskowski RA, Furnham N, Thornton JM. The ramachandran plot and protein structure validation. Biomol Form Func. 2013;62–75. https://doi. org/10.1142/9789814449144_0005
Jiang YL, Zhang JW, Yu WL, et al. Structural and enzymatic characterization of the streptococcal ATP/Diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH. J Biol Chem. 2011;286(41):35906–35914.
Nan J, Brostromer E, Liu XY, Kristensen O, Su XD. Bioinformatics and structural characterization of a hypothetical protein from streptococcus mutans: implication of antibiotic resistance. PloS One. 2009;4(10):e7245. https://doi.org/ 10.1371/journal.pone.0007245
Rabbi MF, Akter SA, Hasan MJ, Amin A. In silico characterization of a hypothetical protein fromShigella DysenteriaeATCC 12039 reveals a pathogenesis-related protein of the Type-Vi secretion system. Bioinf Biol Insights. 2021;15:1–12. https://doi. org/10.1177/11779322211011140
Cozzolino F, Iacobucci I, Monaco V, Monti M. Protein–DNA/Rna interactions: an overview of investigation methods in the -omics era. J Prot Res. 2021;20(6):3018–3030. https://doi.org/10.1021/acs. jproteome.1c00074
Ijaq J, Chandrasekharan M, Poddar R, Bethi N, Sundararajan VS. Annotation and curation of uncharacterized proteins- challenges. Front Genet. 2015;6:e119. https://doi.org/ 10.3389/fgene.2015.00119
Gingras AC, Aebersold R, Raught B. Advances in protein complex analysis using mass spectrometry. J Physiol. 2005;563(1):11–21. https://doi.org/ 10.1113/jphysiol.2004.080440
Fairhead M, Howarth M. Site-Specific biotinylation of purified proteins using BirA. In: Gautier A, Hinner MJ, eds. Site-Specific Protein Labeling. Methods in Molecular Biology. Humana Press; 2014: 171–184. https://doi.org/10.1007/978-1-4939-2272-7_12
Verma V, Kaur C, Grover P, Gupta A, Chaudhary VK. Biotin-tagged proteins: reagents for efficient ELISA-based serodiagnosis and phage display-based affinity selection. PloS One. 2018;13(1):e0191315. https:// doi.org/10.1371/journal.pone.0191315
Wang HZ, Chu ZZ, Chen CC, et al. Recombinant passenger proteins can be conveniently purified by one-step affinity chromatography. PloS One. 2015;10(12):e0143598. https://doi. org/10.1371/journal.pone.0143598
Copyright (c) 2024 Nimra Hanif, Sehrish Arshad, Aqsa , Muhammad Asim, Amna Sadaqat Nadeem, Tanzeel ur Rehman, Nimra shafique, Raees Ahmad khan, Moeez manzor

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










