Impact of Cathepsin L Inhibition in SARS-CoV-2 Infection and Potential Therapeutic Interventions
Abstract
 Abstract Views: 0
Abstract Views: 0
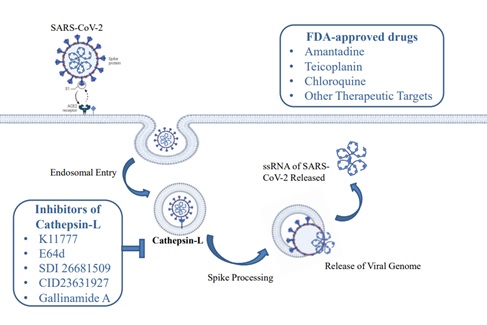
Background. The global COVID-19 pandemic caused by SARS-CoV-2 has prompted the urgent development of effective therapeutic strategies since its emergence in China. Cathepsin L is a lysosomal cysteine protease that plays a pivotal role in the entry of SARS-CoV-2 into the host cell. It follows an endocytic pathway that triggers the fusion of host and viral cell membranes.
Mechanism. Viral RNA is released during this phase and enters the host's cytoplasm through cleavage at S1/S2 or S2′ sites of the Spike glycoprotein of SARS-CoV-2. A study showed K790 as the potential cleavage site for cathepsin L. It is located near the S2′ site on the same loop. Its potential for proteolysis indicates its capacity to induce structural modifications analogous to S2′ cleavage, ultimately activating membrane fusion to allow the entry of the virus. The inhibitors of cathepsin L have emerged as effective drug targets in antiviral therapy.
Conclusion. This study aims to elaborate on the potential role of cathepsin L in SARS-CoV-2 infection during its entry into the host cells and also explores its functional and structural biology. Additionally, it highlights several promising inhibitory compounds including K11777, E64d, SDI 26681509, CID23631927, and Gallinamide A, which are effective in treating the SARS-CoV-2 infection. US Food and Drug Administration (FDA) approved drugs including amantadine, teicoplanin, and chloroquine have the potential to combat the SARS-CoV-2 infection. Understanding the significance of cathepsin L and the use of its inhibitors as therapeutic agents may open new opportunities for developing effective treatments for SARS-CoV-2.
Downloads
References
Lai C-C, Shih T-P, Ko W-C, Tang H-J, Hsueh P-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):e105924. https://doi.org /10.1016/j.ijantimicag.2020.105924
Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12(3):e7423. https://doi.org/10.7759/cureus.7423
Gangavarapu K, Latif AA, Mullen JL, et al. Outbreak. info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat Methods. 2023;20(4):512–522. https://doi.org/10.1038/s41592-023-01769-3
Zahmatkesh S, Sillanpaa M, Rezakhani Y, Wang C. Review of concerned SARS-CoV-2 variants like Alpha (B. 1.1. 7), Beta (B. 1.351), Gamma (P. 1), Delta (B. 1.617. 2), and Omicron (B. 1.1. 529), as well as novel methods for reducing and inactivating SARS-CoV-2 mutants in wastewater treatment facilities. J Hazard Mater Adv. 2022:e100140. https://doi.org/ 10.1016/j.hazadv.2022.100140
Word Health Organization. Number of COVID-19 cases reported to WHO. World Health Organization Web site. https://data.who.int/dashboards/covid19/cases?n=c. Accessed March 3, 2024.
da Rosa Mesquita R, Francelino Silva Junior LC, Santos Santana FM, et al. Clinical manifestations of COVID-19 in the general population: systematic review. Wien Klin Wochenschr. 2021;133(7-8):377–382. https://doi. org/10.1007/s00508-020-01760-4
Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26(7):1017–1032. https://doi.org /10.1038/s41591-020-0968-3
Essalmani R, Jain J, Susan-Resiga D, et al. Distinctive roles of furin and TMPRSS2 in SARS-CoV-2 infectivity. J Virol. 2022;96(8):e00128-22. https://doi.org/ 10.1128/jvi.00128-22
Zhao M-M, Yang W-L, Yang F-Y, et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Sig Transduct Target Ther. 2021;6(1):e134. https://doi.org/ 10.1038/s41392-021-00558-8
Neerukonda S. Therapeutic targets of host factors for potential COVID-19 treatments. In: Jen-Tsung Chen J-T, ed. Bioactive Compounds Against SARS-CoV-2. CRC Press; 2024:23–38.
Kondo Y, Rajapakse S, Ogiwara K. Involvement of cathepsin L in the degradation and degeneration of postovulatory follicle of the medaka ovary. Biol Reproduct. 2023;109(6):904–917. https://doi.org /10.1093/biolre/ioad116
Zhao M-M, Zhu Y, Zhang L, et al. Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanisms for cathepsin L-facilitated viral infection and treatment strategies. Cell Discov. 2022;8(1):e53. https://doi.org/10.1038/s41421-022-00419-w
Liu T, Luo S, Libby P, Shi G-P. Cathepsin L-selective inhibitors: a potentially promising treatment for COVID-19 patients. Pharmacol Therapeut. 2020;213:e107587. https://doi.org/10.1016/j.pharmthera.2020.107587
Gaikwad P. Regulation of Cathepsin L Expression And Activity By Cell Confluence And The Circadian Clock [doctoral dissertation]. Fairborn, United States: Wright State University; 2023.
Reinheckel T, Tholen M. Low‐level lysosomal membrane permeabilization for limited release and sublethal functions of cathepsin proteases in the cytosol and nucleus. FEBS Open Bio. 2022;12(4):694–707. https://doi.org /10.1002/2211-5463.13385
Falke S, Lieske J, Guenther S, et al. Crystal Structure Of Human Cathepsin L With Covalently Bound E-64. Helmholtz Association; 2024. https://doi.org/10.2210/pdb8A4V/pdb
Berdowska I, Matusiewicz M. Cathepsin L, transmembrane peptidase/serine subfamily member 2/4, and other host proteases in COVID-19 pathogenesis–with impact on the gastrointestinal tract. World J Gastroenterol. 2021;27(39): 6590–6600. https://doi.org/10.3748 /wjg.v27.i39.6590
Yadati T, Houben T, Bitorina A, Shiri-Sverdlov R. The ins and outs of cathepsins: physiological function and role in disease management. Cells. 2020;9(7):e1679. https://doi.org/ 10.3390/cells9071679
Yadav R, Chaudhary JK, Jain N, et al. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells. 2021;10(4):e821. https://doi.org/ 10.3390/cells10040821
Gheblawi M, Wang K, Viveiros A, et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circulat Res. 2020;126(10):1456–1474. https://doi.org/10.1161/CIRCRESAHA.120.317015
Bestle D, Heindl MR, Limburg H, et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Allian. 2020;3(9):e202000786. https:// doi.org/10.26508/lsa.202000786
Yang J-K, Zhao M-M, Zhu Y, et al. Novel Sites For Cathepsin L Cleavage in SARS-CoV-2 Spike Guide Treatment Strategies. Research Sequare; 2021. https://doi.org/10 .21203/rs.3.rs-734963/v1
Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScicence. 2020;23(6):e101212. https://doi.org/ 10.1016/j.isci.2020.101212
Zhang Q, Xiang R, Huo S, et al. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct Target Ther. 2021;6(1):e233. https://doi.org/10. 1038/s41392-021-00653-w
Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Nat Acad Sci. 2005;102(33):e11876–11881. https://doi.org/10.1073/pnas.0505577102
Gomes CP, Fernandes DE, Casimiro F, et al. Cathepsin L in COVID-19: from pharmacological pieces of evidence to genetics. Front Cell Infect Microbiol. 2020;10:e589505. https://doi.org/ 10.3389/fcimb.2020.589505
Schroeder JT, Bieneman AP. The S1 subunit of the SARS-CoV-2 spike protein activates human monocytes to produce cytokines linked to COVID-19: relevance to Galectin-3. Front Immunol. 2022;13;831763. https://doi.org/10.3389/fimmu.2022.831763
Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Molecul Cell Biol. 2022;23(1):3–20.
Yu S, Zheng X, Zhou B, et al. SARS-CoV-2 spike engagement of ACE2 primes S2′ site cleavage and fusion initiation. Proc Nat Acad Sci. 2022;119(1):e2111199119. https://doi.org/10.1073/pnas.2111199119
Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):e1620. https://doi.org/10.1038/s41467-020-15562-9
Matveev EV, Ponomarev GV, Kazanov MD. Genome-wide bioinformatics analysis of human protease capacity for proteolytic cleavage of the SARS-CoV-2 spike glycoprotein. Microbiol Spectr. 2024;12(2):e03530-23. https://doi.org/ 10.1128/spectrum.03530-23
Li Q, Zhou S-R, KIM H, Wang H, Zhu J-J, Yang J-K. Discovering novel cathepsin L inhibitors from natural products using artificial intelligence. Comput Struct Biotechnol J. 2024;23:2606–2614. https://doi.org/ 10.1016/j.csbj.2024.06.009
Mellott DM, Tseng C-T, Drelich A, et al. A Cysteine Protease Inhibitor Blocks SARS-CoV-2 Infection of Human and Monkey Cells. bioRxive. https://doi.org/10.1101/2020.10.23.347534
Cannalire R, Stefanelli I, Cerchia C, Beccari AR, Pelliccia S, Summa V. SARS-CoV-2 entry inhibitors: small molecules and peptides targeting virus or host cells. Int J Mol Sci. 2020;21(16):e5707. https://doi.org/ 10.3390/ijms21165707
Pišlar A, Mitrović A, Sabotič J, et al. The role of cysteine peptidases in coronavirus cell entry and replication: the therapeutic potential of cathepsin inhibitors. PLoS Pathog. 2020;16(11):e1009013. https://doi.org /10.1371/journal.ppat.1009013
Mellott DM, Tseng C-T, Drelich A, et al. A clinical-stage cysteine protease inhibitor blocks SARS-CoV-2 infection of human and monkey cells. ACS Chem Biol. 2021;16(4):642–650. https://doi.org/10.1021/acschembio.0c00875
Chang M-C, Chen J-H, Lee H-N, et al. Inducing cathepsin L expression/production, lysosomal activation, and autophagy of human dental pulp cells by dentin bonding agents, camphor quinone, and BisGMA and the related mechanisms. Biomater Adv. 2023;145:e213253. https://doi.org/10.1016/j.bioadv.2022.213253
Acacio Santini Pereira B, Souza-Silva F, Silva-Almeida M, et al. Proteinase inhibitors: a promising drug class for treating leishmaniasis. Curr Drug Targets. 2014;15(12):1121–1131.
Ashhurst AS, Tang AH, Fajtová P, et al. Potent in vitro anti-SARS-CoV-2 activity by gallinamide A and analogs via inhibition of cathepsin L. bioRxiv. 2020:e2020.12.23.424111. https://doi. org/10.1101/2020.12.23.424111
Miller B, Friedman AJ, Choi H, et al. The marine cyanobacterial metabolite gallinamide A is a potent and selective inhibitor of human cathepsin L. J Nat Prod. 2014;77(1):92–99. https://doi.org/10.1021/np400727r
Dana D, Pathak SK. A review of small molecule inhibitors and functional probes of human cathepsin L. Molecules. 2020;25(3):e698. https:// doi.org/10.3390/molecules25030698
Ashhurst AS, Tang AH, Fajtová P, et al. Potent anti-SARS-CoV-2 activity by the natural product gallinamide A and analogs via inhibition of cathepsin L. J Med Chem. 2021;65(4):2956–2970. https://doi.org/10.1021/acs .jmedchem.1c01494
Smieszek SP, Przychodzen BP, Polymeropoulos MH. Amantadine disrupts lysosomal gene expression: a hypothesis for COVID-19 treatment. Int J Antimicrob Agents. 2020;55(6):e106004. https://doi.org/ 10.1016/j.ijantimicag.2020.106004
Toft-Bertelsen TL, Jeppesen MG, Tzortzini E, et al. Amantadine inhibits known and novel ion channels encoded by SARS-CoV-2 in vitro. Commun Biol. 2021;4(1):e1347. https://doi.org/ 10.1038/s42003-021-02866-9
Schütz D, Ruiz-Blanco YB, Münch J, Kirchhoff F, Sanchez-Garcia E, Müller JA. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv Drug Deliver Rev. 2020;167:47–65. https://doi.org/10.1016/j.addr.2020.11.007
Roldan EQ, Biasiotto G, Magro P, Zanella I. The possible mechanisms of action of 4-aminoquinolines (chloroquine/hydroxychloroquine) against Sars-Cov-2 infection (COVID-19): a role for iron homeostasis? Pharmacol Res. 2020;158:e104904. https://doi.org/10.1016/j.phrs.2020.104904
Jaimes-Castelán EG, González-Espinosa C, Magos-Guerrero GA, et al. Drugs and natural products for the treatment of COVID-19 during 2020, the first year of the pandemic. Bol Med Hosp Infant Mex. 2024;81(1):53–72. https://doi.org/10.24875/bmhim.23000016
Babaei F, Mirzababaei M, Nassiri‐Asl M, Hosseinzadeh H. Review of registered clinical trials for the treatment of COVID‐19. Drug Develop Res. 2021;82(4):474–493. https://doi.org/10.1002/ddr.21762
Awadasseid A, Wu Y, Tanaka Y, Zhang W. Effective drugs used to combat SARS-CoV-2 infection and the current status of vaccines. Biomed Pharmacother. 2021;137:e111330. https://doi.org/10.1016/j.biopha.2021.111330
Yousefi B, Valizadeh S, Ghaffari H, Vahedi A, Karbalaei M, Eslami MJ. Global treatments for coronaviruses including COVID‐19. J Cell Phy. 2020;235(12):9133–9142. https://doi.org/10.1002/jcp.29785
Yang W-L, Li Q, Sun J, et al. Potential drug discovery for COVID-19 treatment targeting Cathepsin L using a deep learning-based strategy. Comput Struct Biotechnol J. 2022;20:2442–2454. https://doi.org/ 10.1016/j.csbj.2022.05.023
Falke S, Lieske J, Herrmann A, et al. Structural elucidation and antiviral activity of covalent Cathepsin L inhibitors. J Med Chem. 2024;67(9):7048–7067. https://doi.org /10.1021/acs.jmedchem.3c02351
Milan Bonotto R, Mitrović A, Sosič I, et al. Cathepsin inhibitors nitroxoline and its derivatives inhibit SARS-CoV-2 infection. Antiviral Res. 2023;216:e105655. https://doi.org/10 .1016/j.antiviral.2023.105655
Bobrowski T, Chen L, Eastman RT, et al. Synergistic and antagonistic drug combinations against SARS-CoV-2. Molecul Ther: J Am Soc Gene Ther. 2021;29(2):873–885. https://doi.org/10.1016/j.ymthe.2020.12.016
Copyright (c) 2024 Muteeba Azhar, Asma Irshad, Mehreen Saleem, Tahira Batool

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










