Metabolic and Morphological Analysis of DOF1 Transgenic T2 Wheat Lines under Nitrogen Stress
Abstract
 Abstract Views: 0
Abstract Views: 0
Background. Over the last century, the increased crop production has largely been attributed to rampant input of nitrogen fertilizers causing environmental deterioration. There is a need to engineer crops that require minimal fertilizer input. A transcription factor ‘Triticum aestivum Dof1 (TaDof1)’ is known to improve the nitrogen use efficiency (NUE) of crop plants, by regulating the activity of multiple genes, involved in carbon and nitrogen metabolism, when plants are grown under nitrogen-limiting conditions. Previously, transgenic wheat plants, overexpressing TaDof1, were developed.
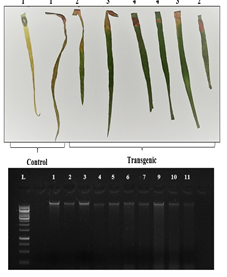
Methods. The main objective of the current study was to compare the T2 generation of six different transgenic wheat lines transformed with TaDof1-namely, F1, G1, G2, G3, G4, and G5-with respect to their metabolic, biochemical, and morphological traits under normal and nitrogen-deficient conditions. BASTA assay and conventional PCR were used to screen out the positive plants. The expression of TaDof1 in aforementioned transgenic lines along with the four genes (Glutamine synthetase, nitrite reductase, phosphoenolpyruvate carboxylase and pyruvate kinase) associated with TaDof1 in carbon and nitrogen metabolism were quantified through RT-PCR and real-time PCR.
Results. The T2 generation of TaDof1 transgenic wheat lines overexpressed the transcription factor TaDof1 along with the other regulated genes. The expression of TaDof1 gene ranged from 0.68-7.61 folds, with the highest fold recorded in line G2. Protein, soluble sugars, phosphorous, chlorophyll, and relative water content were enhanced in almost all transgenic lines.
Conclusion. Overall, galaxy transgenic lines, specifically-G1, showed better metabolic profile as compared to Faisalabad transgenic line under nitrogen stress.
Downloads
References
Borgström P, Bommarco R, Viketoft M, Strengbom J. Below-ground herbivory mitigates biomass loss from above-ground herbivory of nitrogen fertilized plants. Sci Rep. 2020;10:e12752. https://doi.org/ 10.1038/s41598-020-69696-3
Ladha JK, Peoples MB, Reddy PM, et al. Biological nitrogen fixation and prospects for ecological intensification in cereal-based cropping systems. Field Crops Res. 2022;283:e108541. https://doi.org/10.1016/j.fcr.2022.108541
Randive K, Raut T, Jawadand S. An overview of the global fertilizer trends and India’s position in 2020. Miner Econ. 2021;34:371–384. https://doi. org/10.1007/s13563-020-00246-z
Sapkota TB, Bijay-Singh, Takele R. Chapter Five - Improving nitrogen use efficiency and reducing nitrogen surplus through best fertilizer nitrogen management in cereal production: The case of India and China. Adv Agron. 2023;178:233–294. https://doi.org /10.1016/bs.agron.2022.11.006
Sardar MF, Younas F, Farooqi ZU, Li Y. Soil nitrogen dynamics in natural forest ecosystem: a review. Front Fores Global Change. 2023;6:e1144930. https://doi.org/ 10.1038/s41467-024-50803-1
Anas M, Liao F, Verma KK, et al. Fate of nitrogen in agriculture and environment: agronomic, eco-physiological and molecular approaches to improve nitrogen use efficiency. Biol Res. 2020;53:e47. https://doi.org/10.1186/s40659-020-00312-4
Zhang X, Zou T, Lassaletta L, et al. Quantification of global and national nitrogen budgets for crop production. Nat Food. 2021;2:529–540. https://doi.org/10.1038/s43016-021-00318-5
Javed T, Indu I, Singhal RK, et al. Recent advances in agronomic and physio-molecular approaches for improving nitrogen use efficiency in crop plants. Front Plant Sci. 2022;13:e877544. https://doi.org /10.3389/fpls.2022.877544
Charteris AF, Knowles TD, Mead A, Reay MK, Michaelides K, Evershed RP. The differential assimilation of nitrogen fertilizer compounds by soil microorganisms. FEMS Microbiol Lett. 2024;371:efnae041. https://doi. org/10.1093/femsle/fnae041
Zhuo M, Sakuraba Y, Yanagisawa S. Dof1. 7 and NIGT1 transcription factors mediate multilayered transcriptional regulation for different expression patterns of nitrate transporter2 genes under nitrogen deficiency stress. New Phytol. 2024;242(5):2132–2147. https://doi. org/10.1111/nph.19695
Ruta V, Longo C, Lepri A, et al. The DOF transcription factors in seed and seedling development. Plants. 2020;9(2):e218. https://doi.org/10. 3390/plants9020218
Khalid A, Hameed A, Tahir MF. Wheat quality: a review on chemical composition, nutritional attributes, grain anatomy, types, classification, and function of seed storage proteins in bread making quality. Front Nut. 2023;10:e1053196. https://doi.org/10.3389/fnut.2023.1053196
Davies RW, Jakeman PM. Separating the wheat from the chaff: Nutritional value of plant proteins and their potential contribution to human health. Nutrients. 2020;12(8):e2410. https://doi.org/10.3390/nu12082410
Ghafoor I, Habib-Ur-Rahman M, Ali M, et al. Slow-release nitrogen fertilizers enhance growth, yield, NUE in wheat crop and reduce nitrogen losses under an arid environment. Environm Sci Pollut Res. 2021;28(32):43528–43543. https://doi.org/10.1007/s11356-021-13700
Hasnain A, Irfan M, Bashir A, Maqbool A, Malik KA. Transcription factor TaDof1 improves nitrogen and carbon assimilation under low-nitrogen conditions in wheat. Plant Molecul Biol Rep. 2020;38(3):441–451. https://doi.org/10.1007/s11105-020-01208-z
Hoagland DT. Water culture method for growing plants without soil. Circular. 1938;347:1–39.
Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res. 1980;8(19):4321–4326. https://doi.org /10.1093/nar/8.19.4321
Mohsin S, Maqbool A, Ashraf M, Malik KA. Extracellular secretion of phytase from transgenic wheat roots allows utilization of phytate for enhanced phosphorus uptake. Molecul Biotechnol. 2017;59(8):334–342. https://doi.org/10.1007/s12033-017-0020-0
Qiu R, Liu C, Wang Z, Yang Z, Jing Y. Effects of irrigation water salinity on evapotranspiration modified by leaching fractions in hot pepper plants. Sci Rep. 2017;7(1):e7231. https://doi.org/10.1038/s41598-017-07743-2
Kistner C, Matamoros M. RNA isolation using phase extraction and LiCl precipitation. In: Márquez AJ, eds. Lotus Japonicus Handbook. Springer; 2005:123–124. https://doi. org/10.1007/1-4020-3735-x_9
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1-2):248–254. https://doi.org/10. 1016/0003-2697(76)90527-3
Malik CP, Srivastava AK. Text Book of Plant Physiology. Kalyani; 1982.
Arnon DI. Copper enzymes iniisolated chloroplasts. Polyphenoloxidase in Peta Vulgaris. Plant Physiol. 1949;24(1):1–15. https://doi.org /10.1104/pp.24.1.1
Campbell ER, Warsko K, Davidson AM, Campbell WH. Determination of phosphate in soil extracts in the field: a green chemistry enzymatic method. MethodsX. 2015;2:211–218. https:// doi.org/10.1016/j.mex.2015.04.003
Barrs H, Weatherley P. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci. 1962;15(3):e413. https://doi.org /10.1071/bi9620413
Sun L, Di D, Li G, Li Y, Kronzucker HJ. Transcriptome analysis of rice (Oryza sativa L.) in response to ammonium resupply reveals the involvement of phytohormone signaling and the transcription factor OsJAZ9 in reprogramming of nitrogen uptake and metabolism. J Plant Physiol. 2020;246-247:e153137. https://doi.org/10.1016/j.jplph.2020.153137
Neeraja CN, Barbadikar KM, Mangrauthia SK, Rao PR, Subrahmanayam D, Sundaram RM. Genes for NUE in rice: a way forward for molecular breeding and genome editing. Plant Physiol Rep. 2021;26(4):587–599. https://doi.org /10.1007/s40502-021-00632-x
Yu Y, Song T, Wang Y, et al. The wheat WRKY transcription factor TaWRKY1-2D confers drought resistance in transgenic Arabidopsis and wheat (Triticum aestivum L.). Int J Biol Macromol. 2023;226:1203–1217. https://doi.org/10.1016 /j.ijbiomac.2022.11.234
Wulfert S, Schilasky S, Krueger S. Transcriptional and biochemical characterization of cytosolic pyruvate kinases in Arabidopsis thaliana. Plants. 2020;9(3):e353. https://doi.org/10.3390/plants9030353
Domínguez-Figueroa J, Carrillo L, Renau-Morata B, et al. The Arabidopsis transcription factor CDF3 is involved in nitrogen responses and improves nitrogen use efficiency in Tomato. Front Plant Sci. 2020;11:e601558. https://doi.org /10.3389/fpls.2020.601558
Wu D, Li Y, Cao Y, et al. Increased glutamine synthetase by overexpression of TaGS1 improves grain yield and nitrogen use efficiency in rice. Plant Physiol Biochem. 2021;169:259–268. https://doi.org /10.1016/j.plaphy.2021.11.021
Wang D, Xu T, Yin Z, et al. Overexpression of OsMYB305 in rice enhances the nitrogen uptake under low-nitrogen condition. Front Plant Sci. 2020;11:e369. https://doi.org /10.3389/fpls.2020.00369
Copyright (c) 2025 Zulekha Zameer, Kauser A Malik, Asma Maqbool

This work is licensed under a Creative Commons Attribution 4.0 International License.
BSR follows an open-access publishing policy and full text of all published articles is available free, immediately upon publication of an issue. The journal’s contents are published and distributed under the terms of the Creative Commons Attribution 4.0 International (CC-BY 4.0) license. Thus, the work submitted to the journal implies that it is original, unpublished work of the authors (neither published previously nor accepted/under consideration for publication elsewhere). On acceptance of a manuscript for publication, a corresponding author on the behalf of all co-authors of the manuscript will sign and submit a completed the Copyright and Author Consent Form.










